Analysis of histone modification interplay reveals two distinct domains in facultative heterochromatin in Pyricularia oryzae
- PMID: 40695975
- PMCID: PMC12284244
- DOI: 10.1038/s42003-025-08473-2
Analysis of histone modification interplay reveals two distinct domains in facultative heterochromatin in Pyricularia oryzae
Abstract
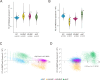
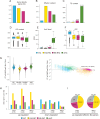
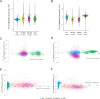
Histone post-translational modifications (PTMs) interact in complex ways to regulate chromatin structure and gene expression. To investigate this interplay, we analyze ChIP-seq and RNA-seq data from knock-out mutants lacking enzymes responsible for H3K4me2/3, H3K9me3, or H3K27me3 in the phytopathogenic fungus Pyricularia oryzae. Loss of specific PTMs alters other PTMs and gene expression in a compartment-specific manner, with distinct effects across H3K4me2-rich euchromatin (EC), H3K27me3-rich facultative heterochromatin (fHC), H3K9me3-rich constitutive heterochromatin (cHC), and centromeres. We identify two distinct fHC subcompartments: K4-fHC, adjacent to EC, and K9-fHC, adjacent to cHC. Both contain poorly conserved genes, but K9-fHC harbors more transposable elements, while K4-fHC is more enriched for genes upregulated during infection, including effector-like genes. H3K27me3 levels in K4-fHC respond to changes in other PTMs, especially H3K9me3, and to environmental conditions. These findings suggest that K4-fHC functions as a reservoir of genes highly responsive to chromatin context and environmental cues.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures





References
-
- Fischle, W., Wang, Y. & Allis, C. D. Histone and chromatin cross-talk. Curr. Opin. Cell Biol.15, 172–183 (2003). - PubMed
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

