IgM+IgD- B cells in human gut-associated lymphoid tissue have memory features and give rise to IgM+ and IgA+ antibody-secreting cells
- PMID: 40695977
- PMCID: PMC12284196
- DOI: 10.1038/s41598-025-11209-1
IgM+IgD- B cells in human gut-associated lymphoid tissue have memory features and give rise to IgM+ and IgA+ antibody-secreting cells
Abstract
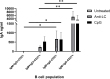
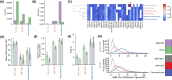
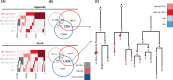
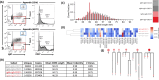
Human IgM+ B cells vary in their surface levels of IgD, with the major circulating population of IgM+IgD+ cells and a minor population (< 5%) of IgM+IgD- cells. In contrast, in gut-associated lymphoid tissue (GALT) derived from individuals undergoing tonsillectomy or appendectomy, IgM+IgD- B cells constitute ~ 30% of B cells. IgM+IgD- cells isolated from both tonsil and appendix lack plasma cell and B1 cell markers, and approximately 50% express the memory marker CD27. Functionally, GALT IgM+IgD- cells spontaneously secrete IgM, and class-switch to IgA in response to both T-dependent and T-independent stimulation ex-vivo. Immune repertoire profiling reveals that GALT IgM+IgD- cells exhibit lower levels of VH4-34 rearrangements, higher levels of somatic hypermutation, shorter CDR3 sequences and greater clonal overlap with switch memory cells than IgM+IgD+ cells. Furthermore, clonal lineage analysis reveals that IgM+IgD- clones can include class-switched sequence variants. These findings suggest a maturational scheme starting from CD27-IgM+IgD+ B cells to CD27+IgM+IgD+, and then to CD27-IgM+IgD-, and finally to CD27+IgM+IgD- B cells. In sum, IgM+IgD- B cells in the mucosa have memory features, give rise to class-switched memory B cells and antibody-secreting cells, and likely contribute significantly to the IgA repertoire in human GALT.
Keywords: B cells; Class-switching; Gut-associated lymphoid tissue (GALT); IgM+ B cells; Memory B cells; Somatic hypermutation (SHM).
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests.
Figures



References
-
- Erika Claud, W. A. W. Chap. 5 - The intestinal microbiota and the microbiome. Gastroenterology and nutrition: neonatology questions and controversies, 73–92, ed. J.N. Richard Polin. W.B. Saunders: Elsevier Inc. (2008).
-
- Morbe, U. M. et al. Human gut-associated lymphoid tissues (GALT); diversity, structure, and function. Mucosal Immunol.14(4), 793–802 (2021). - PubMed
-
- Kagnoff, M. F. Immunology of the intestinal tract. Gastroenterology105(5), 1275–1280 (1993). - PubMed
-
- Spencer, J. & Sollid, L. M. The human intestinal B-cell response. Mucosal Immunol.9(5), 1113–1124 (2016). - PubMed
-
- Michaud, E. et al. Human secretory IgM: an elusive player in mucosal immunity. Trends Immunol.41(2), 141–156 (2020). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

