A cross-sectional molecular epidemiological study of biofilm-producing methicillin-resistant Staphylococcus aureus
- PMID: 40696645
- PMCID: PMC12282711
- DOI: 10.1097/MD.0000000000043346
A cross-sectional molecular epidemiological study of biofilm-producing methicillin-resistant Staphylococcus aureus
Abstract
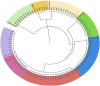
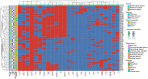
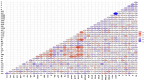
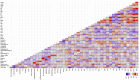
There is growing concern regarding biofilm-producing methicillin-resistant Staphylococcus aureus (MRSA) due to the sudden rise in infection rates and associated morbidity and mortality. Therefore, epidemiological studies, including molecular typing and correlation analysis, are essential for understanding this pathogen. This cross-sectional study investigated epidemiological factors and correlations in MRSA isolates. A total of 300 clinical samples were collected between January and March 2023 from 2 healthcare facilities, including various sample types such as sputum, blood, urine, pus, wound swabs, and other body fluids. This study employed various phenotypic and genotypic methodologies, including adherence assays using standard microtiter plates, the Congo red agar method, antimicrobial resistance and virulence profiling, and multi-locus sequence typing. Among 300 clinical samples from 2 healthcare facilities in Egypt, 94 MRSA isolates were confirmed as biofilm producers. Phylogenetic analysis revealed 8 distinct sequence types (ST8, ST80, ST239, ST15, ST22, ST113, ST398, ST984), found in surgical unit samples across both facilities. Notably, ST22-MRSA was present in all departments, indicating its widespread nature and potential for cross-departmental transmission. ST239-MRSA, the most prevalent strain (22.3%), was found in all departments except burn units. Alarmingly, 95.7% of isolates exhibited multidrug-resistant patterns. However, resistance to vancomycin and imipenem was low among biofilm-producing isolates. The high diversity of MRSA strains suggests multiple sources of infection rather than a single origin. Although most isolates were unrelated, the presence of 2 ST80 isolates in sputum samples from the same unit underscores the importance of targeted infection control within and between hospital areas. ST8-MRSA strains carrying the vanA gene were predominantly identified in body fluid samples, highlighting the need for regular testing in such cases. The diversity of MRSA strains across hospital departments indicates a complex infection landscape with no single source. Although certain genetic markers are linked to specific sequence types, they are not reliable indicators of MRSA clonality. These findings emphasize the need for strict infection control measures and regular testing, particularly for ST8-MRSA in body fluids.
Keywords: MRSA; biofilm; clonality; diversity; sequence types.
Copyright © 2025 the Author(s). Published by Wolters Kluwer Health, Inc.
Conflict of interest statement
The authors have no conflicts of interest to disclose.
Figures

Similar articles
-
Molecular epidemiology, antimicrobial resistance, and virulence characteristics of predominant methicillin-resistant Staphylococcus aureus clones with strong biofilm-producing capability from a tertiary teaching hospital in China.BMC Microbiol. 2025 Aug 15;25(1):510. doi: 10.1186/s12866-025-04258-z. BMC Microbiol. 2025. PMID: 40817043 Free PMC article.
-
Universal versus targeted chlorhexidine and mupirocin decolonisation and clinical and molecular epidemiology of Staphylococcus epidermidis bloodstream infections in patients in intensive care in Scotland, UK: a controlled time-series and longitudinal genotypic study.Lancet Microbe. 2025 Aug;6(8):101118. doi: 10.1016/j.lanmic.2025.101118. Epub 2025 Jun 11. Lancet Microbe. 2025. PMID: 40516572
-
Phenotypic and genotypic characteristics of mecA - positive oxacillin-sensitive Staphylococcus aureus isolated from patients with bloodstream infection in a tertiary hospital in Southern Brazil.Braz J Microbiol. 2024 Sep;55(3):2705-2713. doi: 10.1007/s42770-024-01420-z. Epub 2024 Jun 19. Braz J Microbiol. 2024. PMID: 38896343 Free PMC article.
-
Antibiotic prophylaxis for the prevention of methicillin-resistant Staphylococcus aureus (MRSA) related complications in surgical patients.Cochrane Database Syst Rev. 2013 Aug 19;2013(8):CD010268. doi: 10.1002/14651858.CD010268.pub2. Cochrane Database Syst Rev. 2013. PMID: 23959704 Free PMC article.
-
Antibiotic therapy for the treatment of methicillin-resistant Staphylococcus aureus (MRSA) in non surgical wounds.Cochrane Database Syst Rev. 2013 Nov 18;2013(11):CD010427. doi: 10.1002/14651858.CD010427.pub2. Cochrane Database Syst Rev. 2013. PMID: 24242704 Free PMC article.
References
-
- Türkyilmaz S, Tekbiyik S, Oryasin E, Bozdogan B. Molecular epidemiology and antimicrobial resistance mechanisms of methicillin-resistant Staphylococcus aureus isolated from bovine milk. Zoonoses Public Health. 2010;57:197–203. - PubMed
-
- Graveland H, Duim B, van Duijkeren E, Heederik D, Wagenaar JA. Livestock-associated methicillin-resistant Staphylococcus aureus in animals and humans. Int J Med Microbiol. 2011;301:630–4. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

