Spatially resolved proteomics surveys the chemo-refractory proteins related to high-grade serous ovarian cancer
- PMID: 40697100
- PMCID: PMC12284440
- DOI: 10.1002/ctm2.70422
Spatially resolved proteomics surveys the chemo-refractory proteins related to high-grade serous ovarian cancer
Abstract
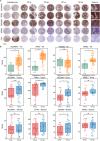
High-grade serous ovarian carcinoma (HGSC) is a lethal malignancy characterized by high incidence, mortality, and chemoresistance. However, its molecular drivers are unknown. In this study, spatially resolved proteomics was applied to 1144 formalin-fixed paraffin-embedded tissue spots obtained by laser capture microdissection from 10 patients with HGSC and divergent carboplatin-paclitaxel (CP) responses. Specific sampling revealed stroma-driven tumour heterogeneity, identifying 642 tumour-specific and 180 stroma-specific proteins, with 505 CP-responsive therapeutic targets. Most of these protein signatures represented previously unreported associations with chemoresistance in HGSCs. Two clinically significant spatial proteomic maps were generated by introducing tumour (TS) and chemical (CS) scores. TS analysis revealed conserved tissue architecture across CP response groups, whereas CS mapping revealed pretreatment metabolic reprogramming (rather than proliferation) as the defining feature of chemo-resistant tumours, challenging current resistance paradigms. Immunohistochemical validation of HGSC tissue microarrays confirmed the spatial proteomic localization of TFRC and PDLIM3, which are linked to tumour progression, while establishing their novel role as chemotherapy resistance biomarkers through this study, with broader predictive potential observed across additional targets in the discovery cohort. This study developed a spatially resolved proteomic framework to enhance the diagnostic and therapeutic strategies for HGSC. KEY POINTS: HGSC intra-tumour heterogeneity is predominantly driven by stroma, as revealed by spatial proteomic compartmentalization (tumour/stroma). Spatial proteomics expands the therapeutic target database, enabling prediction of platinum-based chemotherapy response. Chemo-resistant patients exhibit pre-treatment metabolic activation rather than proliferative signatures. TFRC (iron transport) and PDLIM3 (cytoskeletal remodelling) are spatially validated as chemo-response biomarkers.
Keywords: drug resistance; formalin‐fixed paraffin‐embedded; high‐grade serous ovarian carcinoma; laser capture microdissection; spatial proteomics.
© 2025 The Author(s). Clinical and Translational Medicine published by John Wiley & Sons Australia, Ltd on behalf of Shanghai Institute of Clinical Bioinformatics.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Bell S, McKeeve C, Roxburgh P, Herrington CS. An overview of the molecular pathology of ovarian carcinomas. Diagnos Histopathol. 2024;30:477‐486. doi: 10.1016/j.mpdhp.2024.06.001 - DOI
-
- Zhang Q, Ding J, Wang Y, et al. Tumor microenvironment manipulates chemoresistance in ovarian cancer. Oncol Rep. 2022;47:1‐7. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous
