Genomic and Phenotypic Characterization of Mupirocin-resistant Staphylococcus aureus Clinical Isolates
- PMID: 40698033
- PMCID: PMC12282342
- DOI: 10.1093/ofid/ofaf374
Genomic and Phenotypic Characterization of Mupirocin-resistant Staphylococcus aureus Clinical Isolates
Abstract
Background: Colonization with Staphylococcus aureus is a risk factor for subsequent infection. Decolonization with the topical antibiotic mupirocin is effective and reduces the risk of subsequent S. aureus infection for both methicillin-sensitive and methicillin-resistant (MRSA) strains but may select for mupirocin-resistant isolates.
Methods: We characterized oxacillin and mupirocin susceptibility amongst 384 S. aureus strains isolated from clinical samples isolated in 2017-2023 in Tampa, Florida, spanning strains collected before and after the onset of the coronavirus disease 2019 (COVID-19) pandemic. Whole genome sequencing of bacterial isolates was conducted in parallel and correlated with drug susceptibility profiles.
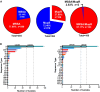
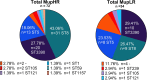
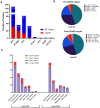
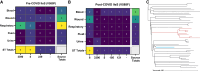
Results: Mupirocin resistance (MupR) was nearly exclusively present in MRSA strains (103/106, 97.1% of MupR; 103/299, 34.4% of MRSA). Although our hospital protocol for decolonization shifted to povidone iodine in the post-COVID period, the overall prevalence of MupR did not change in pre-COVID and post-COVID samples (28.9% vs 26%). Genotype correlated with antibiotic susceptibility with low-level MupR, linked to mutations in ileS and high-level MupR, linked to the presence of mupA. Genome analysis revealed that most MupR strains fell into 3 sequence types (ST) falling into 2 major clonal complexes (CC): CC8 ST8 (including community-associated MRSA strains USA300 and USA500), CC5 ST5 (associated with healthcare-associated MRSA such as USA100), and CC5 ST3390. ST3390 isolates had the highest prevalence of MupR (30/36 83%; high-level MupR 20/36 55.6%; low-level MupR 10/36 27.8%).
Conclusions: Mupirocin resistance was prevalent in our hospital MRSA strains. We also found evidence for emergence and persistence of ST3390 MRSA-MupR strains in Florida.
Keywords: MRSA; antimicrobial resistance; decolonization; mupirocin; staphylococcus aureus.
© The Author(s) 2025. Published by Oxford University Press on behalf of Infectious Diseases Society of America.
Conflict of interest statement
Potential conflicts of interest. K.K. is a member of the Editorial Board of the Sanford Guide, has consulted for Shionogi and Regeneron, and has received clinical research grants from Regeneron, Astra Zeneca, Pfizer, and the NIH. All other authors report no potential conflicts.
Figures
Update of
-
Genomic and Phenotypic Characterization of Mupirocin Resistant Staphylococcus aureus Clinical Isolates.bioRxiv [Preprint]. 2025 Jun 2:2025.04.16.648996. doi: 10.1101/2025.04.16.648996. bioRxiv. 2025. Update in: Open Forum Infect Dis. 2025 Jul 15;12(7):ofaf374. doi: 10.1093/ofid/ofaf374. PMID: 40501773 Free PMC article. Updated. Preprint.
References
Grants and funding
LinkOut - more resources
Full Text Sources

