Immuno-informatics analyses of important esophageal cancer associated viruses for multi-epitope vaccine design
- PMID: 40698087
- PMCID: PMC12279721
- DOI: 10.3389/fimmu.2025.1587224
Immuno-informatics analyses of important esophageal cancer associated viruses for multi-epitope vaccine design
Abstract
Introduction: Esophageal cancer (EC) is a highly lethal malignancy characterized by the uncontrolled proliferation of cancerous cells within the esophagus. Despite recent advancements in therapeutic strategies, the prognosis remains poor, underscoring the urgent need for novel preventive and therapeutic approaches. Notably, several oncogenic viruses have been implicated in EC pathogenesis, prompting the exploration of epitope-based vaccines through immunoinformatics.
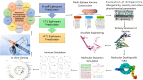
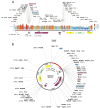
Methods: Using immunoinformatics and bioinformatics approaches, we designed a novel multi-epitope vaccine targeting viral agents associated with EC. Protein sequences of ten viral candidates were retrieved from the UniProt database and evaluated for antigenicity using the VaxiJen server. Five highly antigenic proteins derived from Human Cytomegalovirus (HCMV), Human Papillomavirus (HPV), Human Herpesvirus 8 (HHV-8), Human Immunodeficiency Virus (HIV), and Epstein-Barr Virus (EBV) were selected. T cell (CTL and HTL) and B cell (LBL) epitopes were predicted and screened for immunogenicity, allergenicity, and toxicity. The final vaccine construct incorporated β-defensin as an adjuvant and included 3 HTL, 8 CTL, and 8 LBL epitopes. Molecular docking and molecular dynamics (MD) simulations were conducted to assess the binding affinity of the vaccine with Toll-like receptor 3 (TLR3). In silico cloning was also performed using the pET-28a(+) vector in Escherichia coli strain K12.
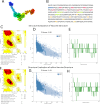
Results: The designed vaccine was found to be antigenic, non-allergenic, and non-toxic. Molecular docking revealed strong binding affinity between the vaccine construct and TLR3, which was further supported by MD simulation results indicating stable complex formation. Codon optimization and in silico cloning confirmed the high expression potential of the vaccine in the E. coli expression system.
Discussion: The in silico analyses suggest that the developed multi-epitope vaccine construct is a promising candidate for preventing EC associated with viral infections. While these findings are encouraging, further experimental validation through in vitro and in vivo studies is essential to confirm the vaccine's safety, immunogenicity, and protective efficacy.
Keywords: Epstein-Barr virus (EBV); esophageal cancer; human cytomegalovirus (hcmv); human papillomavirus (HPV); molecular docking simulation; multi-epitope vaccine.
Copyright © 2025 Shehri.
Conflict of interest statement
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures






References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

