Upregulation of ARHGAP18 by miR-613 Inhibits Cigarette Smoke Extract-Induced Apoptosis and Epithelial-Mesenchymal Transition in Bronchial Epithelial Cells
- PMID: 40698118
- PMCID: PMC12282545
- DOI: 10.2147/COPD.S524723
Upregulation of ARHGAP18 by miR-613 Inhibits Cigarette Smoke Extract-Induced Apoptosis and Epithelial-Mesenchymal Transition in Bronchial Epithelial Cells
Abstract
Objective: Chronic Obstructive Pulmonary Disease (COPD) is a major chronic respiratory disease affecting human health worldwide. However, there is still a lack of effective drugs for treating COPD. This study is intended to explore the function and molecular mechanism of ARHGAP18 and miR-613 in COPD pathogenesis.
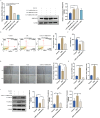
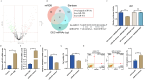
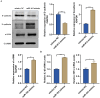
Methods: We initially identified the marker gene closely related to epithelial dysfunction in COPD by integrating bioinformatic analyses. ARHGAP18 expression in CSE-induced bronchial epithelial cells (BEAS-2B) was detected by qRT-PCR. Besides, ARHGAP18 levels were modulated by lentivirus-mediated overexpression. Thereafter, cell variability, apoptosis, and migration were detected by CCK8, flow cytometry, and wound healing assay. IL-1β and TNF-α levels were examined by qRT-PCR. Epithelial-mesenchymal transition (EMT)-associated proteins were determined by Western blotting. The function of miR-613 in COPD was further detected. Functional rescue experiments were performed to determine the mechanism of ARHGAP18 in COPD.
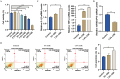
Results: Our study identified ARHGAP18 as the key gene associated with epithelial dysfunction in COPD. ARHGAP18 was downregulated in CSE-induced BEAS-2B cells. Overexpression of ARHGAP18 inhibited cell apoptosis of BEAS-2B cells and enhanced their proliferation and migration. Besides, ARHGAP18 overexpression reduced IL-1 β and TNF-α levels, enhanced E-cadherin expression, and suppressed Vimentin and N-cadherin expression. In contrast, miR-613 mimics exerted opposite effects. Furthermore, downregulation of ARHGAP1, mediated by miR-613 inhibitor promoted cell apoptosis and EMT of CSE-induced BEAS-2B cells, suggesting a regulatory role of miR-613 in COPD pathogenesis.
Conclusion: These findings highlight miR-613/ARHGAP18 axis as a critical regulator of epithelial dysfunction in COPD, offering a potential therapeutic target to counteract apoptosis, inflammation, and airway remodeling.
Keywords: ARHGAP18; apoptosis; chronic obstructive pulmonary disease; epithelial-mesenchymal transition; miR-613.
© 2025 Fu et al.
Conflict of interest statement
There are no conflicts of interest related to this study.
Figures


Similar articles
-
hUC-MSC-derived extracellular vesicles protect lung tissue from cigarette smoke-induced injury by upregulating the METTL3-mediated m6A modification.Am J Physiol Cell Physiol. 2025 Aug 1;329(2):C426-C439. doi: 10.1152/ajpcell.00222.2025. Epub 2025 Jun 23. Am J Physiol Cell Physiol. 2025. PMID: 40549554
-
Cigarette Smoke Extract-Induced Necroptosis Causes Mitochondrial DNA Release and Inflammation of Bronchial Epithelial Cells.Int J Chron Obstruct Pulmon Dis. 2025 Aug 1;20:2685-2695. doi: 10.2147/COPD.S523610. eCollection 2025. Int J Chron Obstruct Pulmon Dis. 2025. PMID: 40765678 Free PMC article.
-
GLUT3-mediated cigarette smoke-induced epithelial-mesenchymal transition in chronic obstructive pulmonary disease through the NF-kB/ZEB1 pathway.Respir Res. 2024 Apr 9;25(1):158. doi: 10.1186/s12931-024-02785-3. Respir Res. 2024. PMID: 38594707 Free PMC article.
-
Computer and mobile technology interventions for self-management in chronic obstructive pulmonary disease.Cochrane Database Syst Rev. 2017 May 23;5(5):CD011425. doi: 10.1002/14651858.CD011425.pub2. Cochrane Database Syst Rev. 2017. PMID: 28535331 Free PMC article.
-
Self-management interventions for people with chronic obstructive pulmonary disease.Cochrane Database Syst Rev. 2022 Jan 10;1(1):CD002990. doi: 10.1002/14651858.CD002990.pub4. Cochrane Database Syst Rev. 2022. PMID: 35001366 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

