Unraveling the therapeutic landscape of approved non-peptide macrocycles
- PMID: 40698129
- PMCID: PMC12278416
- DOI: 10.1016/j.apsb.2025.04.021
Unraveling the therapeutic landscape of approved non-peptide macrocycles
Abstract
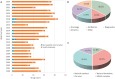
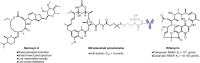
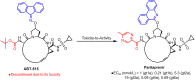
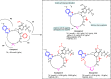
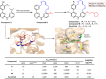
Non-peptide macrocyclic drugs possess unique structural advantages that allow them to target various biomolecules of interest and thus show therapeutic potential against various diseases such as cancer, infectious diseases, etc. This review article examines 34 non-peptide macrocyclic drugs approved between 2000 and 2024, with a particular focus on the optimization process of representative macrocyclic drugs such as natural macrocycles, natural product-inspired macrocycles, and de novo-designed macrocycles. We discuss their structural characteristics, highlighting how conformational rigidity and enhanced target specificity contribute to their efficacy. Design details of these new macrocyclic drugs are illustrated through successful examples, offering insights for optimizing macrocycles. Of note, macrocyclization of U-shaped lead structures represents a novel molecular skeleton editing strategy in de novo macrocycle drug design.
Keywords: Drug discovery; Macrocycles; Macrocyclization; Molecular editing; Non-peptides.
© 2025 The Authors.
Conflict of interest statement
The authors have no conflicts of interest to declare.
Figures













References
-
- Giordanetto F., Kihlberg J. Macrocyclic drugs and clinical candidates: what can medicinal chemists learn from their properties? J Med Chem. 2014;57:278–295. - PubMed
-
- Hann M.M., Keseru G.M. Finding the sweet spot: the role of nature and nurture in medicinal chemistry. Nat Rev Drug Discov. 2012;11:355–365. - PubMed
-
- Zhang G., Zhang J., Gao Y., Li Y., Li Y. Strategies for targeting undruggable targets. Expet Opin Drug Discov. 2022;17:55–69. - PubMed
-
- You L., An R., Liang K., Cui B., Wang X. Macrocyclic compounds: emerging opportunities for current drug discovery. Curr Pharm Des. 2016;22:4086–4093. - PubMed
-
- Driggers E.M., Hale S.P., Lee J., Terrett N.K. The exploration of macrocycles for drug discovery—an underexploited structural class. Nat Rev Drug Discov. 2008;7:608–624. - PubMed
Publication types
LinkOut - more resources
Full Text Sources

