Phase variation in Mannheimia haemolytica challenges the static genome paradigm
- PMID: 40698827
- PMCID: PMC12403840
- DOI: 10.1128/spectrum.00010-25
Phase variation in Mannheimia haemolytica challenges the static genome paradigm
Abstract
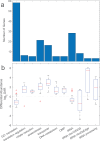
Mannheimia haemolytica is a key agent in bovine respiratory disease (BRD), driving antibiotic use in feedyard cattle. As a facultative anaerobe commonly found in the upper respiratory tracts of cattle, its role under anaerobic conditions in BRD has not been extensively studied despite the known importance of anaerobiosis in human respiratory infections. Utilizing a combined omics approach, we refuted the null hypothesis that the M. haemolytica genome sequence is independent of the environment. This finding provides the rationale for research exploring how anaerobiosis-driven genome plasticity might contribute to a transition from a commensal to a virulent state. Genome sequencing of colony morphology variants from aerobic and anaerobic cultures revealed phase variation via homologous recombination between ribosomal RNA (rRNA) operons and slipped strand mispairing at simple sequence repeats (SSRs), frameshifting genes "on" and "off." Homologous recombination was exclusive to anaerobic conditions. SSR length variation in a DNA methyltransferase gene, targeting 5'-GAC
Keywords: DNA methylation; aerobiosis; anaerobiosis; bacterial infection; cattle; homologous recombination; hypoxia; nitrate reduction; persistence; phase variation; respiratory disease; ruminants; simple sequence repeat; slipped strand mispairing; tandem repeat.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

