Subtypes and proliferation patterns of small intestine neuroendocrine tumors revealed by single-cell RNA sequencing
- PMID: 40699214
- PMCID: PMC12286602
- DOI: 10.7554/eLife.101153
Subtypes and proliferation patterns of small intestine neuroendocrine tumors revealed by single-cell RNA sequencing
Abstract
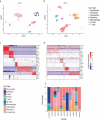
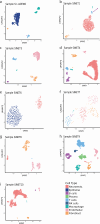
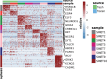
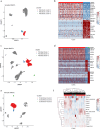
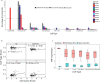
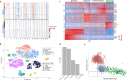
Neuroendocrine tumors (NETs) occur primarily in the small intestine, lung, and pancreas. Due to their rarity compared to other malignancies in these organs, their complex biology remains poorly understood, including their oncogenesis, tumor composition, and the intriguing phenomena of mixed neuroendocrine non-neuroendocrine neoplasms (MiNEN). Here, we profiled ten low-grade small intestine NET (SiNET) samples as well as one mixed lung tumor by single-cell or single-nuclei RNA-seq. We find that SiNETs are largely separated into two distinct subtypes, in which the neuroendocrine cells upregulate epithelial or neuronal markers, respectively. Surprisingly, in both subtypes, the neuroendocrine cells are largely non-proliferative while higher proliferation is observed in multiple non-malignant cell types. Specifically, B and plasma cells are highly proliferative in the epithelial-like SiNET subtype, potentially reflecting the outcome of high Migration Inhibitory Factor (MIF) expression in those tumors, which may constitute a relevant target. Finally, our analysis of a mixed lung neuroendocrine tumor identifies a population of putative progenitor cells that may give rise to both neuroendocrine and non-neuroendocrine (squamous) cells, potentially explaining the origin of the mixed histology. Taken together, our results provide important insights and hypotheses regarding the biology of neuroendocrine neoplasms.
Keywords: cancer biology; cell cycle; human; neuroendocrine tumors; single-cell RNA-seq; small intestine; tumor heterogeneity.
© 2024, Somech, Halder, Spitzer et al.
Conflict of interest statement
ES, DH, AS, CB, MT, RH, MB, AT No competing interests declared, IT advisory board member of Immunitas Therapeutics, and a co-founder and advisory board member of Cellyrix Therapeutics
Figures




Update of
- doi: 10.1101/2024.07.29.605642
- doi: 10.7554/eLife.101153.1
- doi: 10.7554/eLife.101153.2
References
-
- Alvarez MJ, Subramaniam PS, Tang LH, Grunn A, Aburi M, Rieckhof G, Komissarova EV, Hagan EA, Bodei L, Clemons PA, Dela Cruz FS, Dhall D, Diolaiti D, Fraker DA, Ghavami A, Kaemmerer D, Karan C, Kidd M, Kim KM, Kim HC, Kunju LP, Langel Ü, Li Z, Lee J, Li H, LiVolsi V, Pfragner R, Rainey AR, Realubit RB, Remotti H, Regberg J, Roses R, Rustgi A, Sepulveda AR, Serra S, Shi C, Yuan X, Barberis M, Bergamaschi R, Chinnaiyan AM, Detre T, Ezzat S, Frilling A, Hommann M, Jaeger D, Kim MK, Knudsen BS, Kung AL, Leahy E, Metz DC, Milsom JW, Park YS, Reidy-Lagunes D, Schreiber S, Washington K, Wiedenmann B, Modlin I, Califano A. A precision oncology approach to the pharmacological targeting of mechanistic dependencies in neuroendocrine tumors. Nature Genetics. 2018;50:979–989. doi: 10.1038/s41588-018-0138-4. - DOI - PMC - PubMed
-
- Andersson E, Arvidsson Y, Swärd C, Hofving T, Wängberg B, Kristiansson E, Nilsson O. Expression profiling of small intestinal neuroendocrine tumors identifies subgroups with clinical relevance, prognostic markers and therapeutic targets. Modern Pathology. 2016;29:616–629. doi: 10.1038/modpathol.2016.48. - DOI - PubMed
MeSH terms
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Miscellaneous

