Mapping Small Extracellular Vesicle Secretion Potential in Healthy Human Gingiva Using Spatial Transcriptomics
- PMID: 40699655
- PMCID: PMC12026306
- DOI: 10.3390/cimb47040256
Mapping Small Extracellular Vesicle Secretion Potential in Healthy Human Gingiva Using Spatial Transcriptomics
Abstract
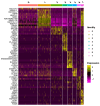
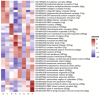
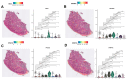
Regenerative processes occur at various levels in all organisms, yet their complexity continues to raise new questions about their mechanisms. It has been demonstrated that small extracellular vesicles (sEVs), secreted by all cells and influencing their function, play a significant role in regeneration. In the context of regenerative processes, oral mucosal tissues consistently receive interest, as they are among the most rapidly healing tissues in the human body. In this study, we utilized spatial transcriptomics to map gene expression to specific spatial locations within the gingiva tissue section, using publicly available transcriptomic data. This analysis revealed new insights into this tissue and the biogenesis of sEVs within it. The identified clusters encompassed two main regions-the epithelium and lamina propria-as well as minor niches within them. Using Gene Ontology (GO) analysis, we identified two clusters most enriched in extracellular vesicle-related GO processes. These included the superficial and deeper layers of the sulcular epithelium, one of the most peripheral regions of the gingiva. Of the 43 genes identified in the literature as having a potential or documented role in sEVs biogenesis, 12 were selected for further analysis. MUC1, SDCBP2, and VPS37B showed clear specificity and the highest expression in the superficial layer of the sulcular epithelium. CHMP4C also exhibited high expression in this layer, though its levels were comparable to the outer layer of the oral epithelium. Other well-established sEVs marker genes, such as ANXA2, CD9, CD63, CD81, FLOT1, RAB22A, RAB27B, and RAB5A, were also expressed in the examined tissue; however, their expression was not specifically exclusive to the sulcular epithelium. Our study is the first to perform a meta-analysis of available gingival transcriptomic data in the specific context of sEVs biogenesis. The presented data and conclusions provide new insights into the role of different structures within healthy human gingiva and shed new light on both known and potential markers of sEVs biogenesis. These findings may contribute to the development of regeneration-targeted research, especially on oral tissues.
Keywords: gingiva; oral mucosa; regeneration; small extracellular vesicles; spatial transcriptomics.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


Similar articles
-
The Black Book of Psychotropic Dosing and Monitoring.Psychopharmacol Bull. 2024 Jul 8;54(3):8-59. Psychopharmacol Bull. 2024. PMID: 38993656 Free PMC article. Review.
-
Sexual Harassment and Prevention Training.2024 Mar 29. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2024 Mar 29. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 36508513 Free Books & Documents.
-
Falls prevention interventions for community-dwelling older adults: systematic review and meta-analysis of benefits, harms, and patient values and preferences.Syst Rev. 2024 Nov 26;13(1):289. doi: 10.1186/s13643-024-02681-3. Syst Rev. 2024. PMID: 39593159 Free PMC article.
-
Signs and symptoms to determine if a patient presenting in primary care or hospital outpatient settings has COVID-19.Cochrane Database Syst Rev. 2022 May 20;5(5):CD013665. doi: 10.1002/14651858.CD013665.pub3. Cochrane Database Syst Rev. 2022. PMID: 35593186 Free PMC article.
-
Drugs for preventing postoperative nausea and vomiting in adults after general anaesthesia: a network meta-analysis.Cochrane Database Syst Rev. 2020 Oct 19;10(10):CD012859. doi: 10.1002/14651858.CD012859.pub2. Cochrane Database Syst Rev. 2020. PMID: 33075160 Free PMC article.
References
-
- Prabhu S.R. Textbook of General Pathology for Dental Students. Springer Nature; Berlin/Heidelberg, Germany: 2023. Healing: Tissue Regeneration and Repair; pp. 49–56. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

