Reconstructing Dynamic Gene Regulatory Networks Using f-Divergence from Time-Series scRNA-Seq Data
- PMID: 40699807
- PMCID: PMC12192124
- DOI: 10.3390/cimb47060408
Reconstructing Dynamic Gene Regulatory Networks Using f-Divergence from Time-Series scRNA-Seq Data
Abstract
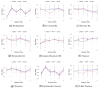
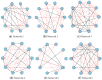
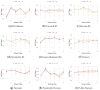
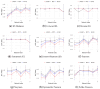
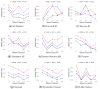
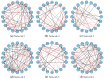
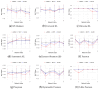
Inferring time-varying gene regulatory networks from time-series single-cell RNA sequencing (scRNA-seq) data remains a challenging task. The existing methods have notable limitations as most are either designed for reconstructing time-varying networks from bulk microarray data or constrained to inferring stationary networks from scRNA-seq data, failing to capture the dynamic regulatory changes at the single-cell level. Furthermore, scRNA-seq data present unique challenges, including sparsity, dropout events, and the need to account for heterogeneity across individual cells. These challenges complicate the accurate capture of gene regulatory network dynamics over time. In this work, we propose a novel f-divergence-based dynamic gene regulatory network inference method (f-DyGRN), which applies f-divergence to quantify the temporal variations in gene expression across individual single cells. Our approach integrates a first-order Granger causality model with various regularization techniques and partial correlation analysis to reconstruct gene regulatory networks from scRNA-seq data. To infer dynamic regulatory networks at different stages, we employ a moving window strategy, which allows for the capture of dynamic changes in gene interactions over time. We applied this method to analyze both simulated and real scRNA-seq data from THP-1 human myeloid monocytic leukemia cells, comparing its performance with the existing approaches. Our results demonstrate that f-DyGRN, when equipped with a suitable f-divergence measure, outperforms most of the existing methods in reconstructing dynamic regulatory networks from time-series scRNA-seq data.
Keywords: Granger causality; f-divergence; gene regulatory network; regularization; single-cell RNA sequencing; time-series data; time-varying network.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
Similar articles
-
MuDCoD: multi-subject community detection in personalized dynamic gene networks from single-cell RNA sequencing.Bioinformatics. 2023 Oct 3;39(10):btad592. doi: 10.1093/bioinformatics/btad592. Bioinformatics. 2023. PMID: 37740957 Free PMC article.
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2021 Apr 19;4(4):CD011535. doi: 10.1002/14651858.CD011535.pub4. Cochrane Database Syst Rev. 2021. Update in: Cochrane Database Syst Rev. 2022 May 23;5:CD011535. doi: 10.1002/14651858.CD011535.pub5. PMID: 33871055 Free PMC article. Updated.
-
scRegulate: Single-Cell Regulatory-Embedded Variational Inference of Transcription Factor Activity from Gene Expression.bioRxiv [Preprint]. 2025 May 5:2025.04.17.649372. doi: 10.1101/2025.04.17.649372. bioRxiv. 2025. PMID: 40654959 Free PMC article. Preprint.
-
Soft graph clustering for single-cell RNA sequencing data.BMC Bioinformatics. 2025 Jul 25;26(1):195. doi: 10.1186/s12859-025-06231-z. BMC Bioinformatics. 2025. PMID: 40713495 Free PMC article.
-
Surgical interventions for treating extracapsular hip fractures in older adults: a network meta-analysis.Cochrane Database Syst Rev. 2022 Feb 10;2(2):CD013405. doi: 10.1002/14651858.CD013405.pub2. Cochrane Database Syst Rev. 2022. PMID: 35142366 Free PMC article.
References
Grants and funding
LinkOut - more resources
Full Text Sources

