Selective Dual Inhibition of TNKS1 and CDK8 by TCS9725 Attenuates STAT1/β-Catenin/TGFβ1 Signaling in Renal Cancer
- PMID: 40699862
- PMCID: PMC12192500
- DOI: 10.3390/cimb47060463
Selective Dual Inhibition of TNKS1 and CDK8 by TCS9725 Attenuates STAT1/β-Catenin/TGFβ1 Signaling in Renal Cancer
Abstract
Background: Tankyrase (TNKS1) regulates the WNT/β-catenin pathway, while CDK8 is a transcriptional regulator overexpressed in renal cell carcinoma (RCC). This study aims to identify novel dual inhibitors of tankyrase and Cyclin-dependent kinase 8 (CDK8), utilizing bioinformatics and in vitro methods and to assess their efficiency in renal cancer cells.
Methods: To identify leads, the ChemBridge library was screening using high-throughput virtual screening (HTVS), which was followed by protein-ligand interaction analysis, Molecular Dynamics (MD) simulation, and Gibbs binding free energy estimation. A-498, Caki-1, and HK-2 cells were employed to validate in vitro efficacy.
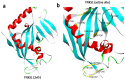
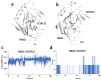
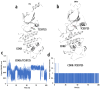
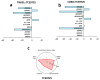
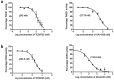
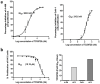
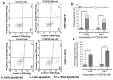
Results: TCS9725 was discovered by HTVS with binding affinities of -8.1 kcal/mol and -8.2 kcal/mol for TNKS1 and CDK8, respectively. TCS9725 had robust binding interactions with root mean square deviation values of 0.00 nm. The ΔG binding estimate was -27.45 for TNKS1 and -27.88 for CDK8, respectively. ADME predictions favored specific small-molecule inhibition profiles. TCS9725 reduced TNKS1 and CDK8 activities with IC50s of 243 nM and 403.6 nM, respectively. The compound efficiently inhibited the growth of A-498 and Caki-1 cells with GI50 values of 385.9 nM and 243.6 nM, respectively, with high selectivity compared to the non-cancerous kidney cells. TCS9725 decreased STAT1 and β-catenin positivity in A-498 and Caki-1 cells. The compound induced apoptosis and reduced TGFβ-stimulated trans-endothelial migration and p-smad2/3 signaling in both RCC cells.
Conclusions: This work provides valuable insights into the therapeutic potential of TCS9725, a dual inhibitor of TNKS1 and CDK8. Further developments of this molecule could lead to new and effective treatments for this devastating disease.
Keywords: CDK8; TNKS1; WNT/β-catenin; dual inhibition; renal cancer.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures




References
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

