Meta-Analysis of Gene Expression in Bulk-Processed Post-Mortem Spinal Cord from ALS Patients and Normal Controls
- PMID: 40700130
- PMCID: PMC12286074
- DOI: 10.3390/neurosci6030065
Meta-Analysis of Gene Expression in Bulk-Processed Post-Mortem Spinal Cord from ALS Patients and Normal Controls
Abstract
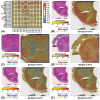
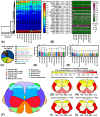
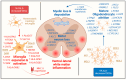
Amyotrophic lateral sclerosis (ALS) is characterized by upper and lower motor neuron failure and poor prognosis. This study performed a meta-analysis of gene expression datasets that compared bulk-processed post-mortem spinal cord from ALS and control (CTL) patients. The analysis included 569 samples (454 ALS, 115 CTL) from 348 individuals (262 ALS, 86 CTL). Patterns of differential expression bias, related to mRNA abundance, gene length and GC content, were discernable from individual studies but attenuated by meta-analysis. A total of 213 differentially expressed genes (DEGs) were identified (144 ALS-increased, 69 ALS-decreased). ALS-increased DEGs were most highly expressed by microglia and associated with MHC class II, immune response and leukocyte activation. ALS-decreased DEGs were abundantly expressed by mature oligodendrocytes (e.g., the MOL5 phenotype) and associated with myelin production, plasma membrane and sterol metabolism. Comparison to spatial transcriptomics data showed that DEGs were prominently expressed in white matter, with increased DEG expression strongest in the ventral/lateral white matter. These results highlight white matter as the spinal cord region most strongly associated with the shifts in mRNA abundance observed in bulk-processed tissues. These shifts can be explained by attrition of mature oligodendrocytes and an ALS-emergent microglia phenotype that is partly shared among neurodegenerative conditions.
Keywords: RNA-seq; microglia; motor neuron disease; neuromuscular; oligodendrocyte; spatial transcriptomics.
Conflict of interest statement
The author declares no conflicts of interest.
Figures






Similar articles
-
Mechanical ventilation for amyotrophic lateral sclerosis/motor neuron disease.Cochrane Database Syst Rev. 2017 Oct 6;10(10):CD004427. doi: 10.1002/14651858.CD004427.pub4. Cochrane Database Syst Rev. 2017. PMID: 28982219 Free PMC article.
-
Behavioral interventions to reduce risk for sexual transmission of HIV among men who have sex with men.Cochrane Database Syst Rev. 2008 Jul 16;(3):CD001230. doi: 10.1002/14651858.CD001230.pub2. Cochrane Database Syst Rev. 2008. PMID: 18646068
-
Translating state-of-the-art spinal cord MRI techniques to clinical use: A systematic review of clinical studies utilizing DTI, MT, MWF, MRS, and fMRI.Neuroimage Clin. 2015 Dec 4;10:192-238. doi: 10.1016/j.nicl.2015.11.019. eCollection 2016. Neuroimage Clin. 2015. PMID: 26862478 Free PMC article.
-
Gamma aminobutyric acid (GABA) modulators for amyotrophic lateral sclerosis/motor neuron disease.Cochrane Database Syst Rev. 2017 Jan 9;1(1):CD006049. doi: 10.1002/14651858.CD006049.pub2. Cochrane Database Syst Rev. 2017. PMID: 28067943 Free PMC article.
-
Management of faecal incontinence and constipation in adults with central neurological diseases.Cochrane Database Syst Rev. 2014 Jan 13;2014(1):CD002115. doi: 10.1002/14651858.CD002115.pub5. Cochrane Database Syst Rev. 2014. Update in: Cochrane Database Syst Rev. 2024 Oct 29;10:CD002115. doi: 10.1002/14651858.CD002115.pub6. PMID: 24420006 Free PMC article. Updated.
References
-
- Wu J.Y., Ye S., Liu X.Y., Xu Y.S., Fan D.S. The burden of upper motor neuron involvement is correlated with the bilateral limb involvement interval in patients with amyotrophic lateral sclerosis: A retrospective observational study. Neural Regen. Res. 2025;20:1505–1512. doi: 10.4103/NRR.NRR-D-23-01359. - DOI - PMC - PubMed
-
- Calderón-Garcidueñas L., Stommel E.W., Torres-Jardón R., Hernández-Luna J., Aiello-Mora M., González-Maciel A., Reynoso-Robles R., Pérez-Guillé B., Silva-Pereyra H.G., Tehuacanero-Cuapa S., et al. Alzheimer and Parkinson diseases, frontotemporal lobar degeneration and amyotrophic lateral sclerosis overlapping neuropathology start in the first two decades of life in pollution exposed urbanites and brain ultrafine particulate matter and industrial nanoparticles, including Fe, Ti, Al, V, Ni, Hg, Co, Cu, Zn, Ag, Pt, Ce, La, Pr and W are key players. Metropolitan Mexico City health crisis is in progress. Front. Hum. Neurosci. 2024;17:1297467. doi: 10.3389/fnhum.2023.1297467. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

