Comparative Label-Based Proteomics of Venoms from Echis ocellatus, Naja nigricollis, and Bitis arietans
- PMID: 40700275
- PMCID: PMC12286013
- DOI: 10.3390/proteomes13030031
Comparative Label-Based Proteomics of Venoms from Echis ocellatus, Naja nigricollis, and Bitis arietans
Abstract
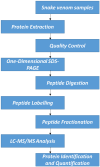
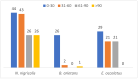
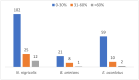
Background: Snake envenomation is a major public health issue in Nigeria, primarily due to bites from Echis ocellatus, Naja nigricollis, and Bitis arietans. Understanding their venom composition is essential for effective antivenom development. This study characterizes and compares the venom proteomes of these snakes using iTRAQ-based proteomics, focusing on key toxin families and their relative abundances. Methods: Venom samples were ethically collected from adult snakes, pooled by species, lyophilized, and stored for proteomic analysis. Proteins were extracted, digested with trypsin, and labeled with iTRAQ. Peptides were analyzed via mass spectrometry, and data were processed using Mascot and IQuant for protein identification and quantification. Results:E. ocellatus and B. arietans venoms had similar profiles, rich in C-type lectins, serine proteases, and phospholipase A2s. These comprised 17%, 11%, and 5% in E. ocellatus and 47%, 10%, and 7% in B. arietans, with metalloproteinases dominating both (53% and 47%). In N. nigricollis, three-finger toxins (9%) were most abundant, followed by metalloproteinases (3%). All species shared four core protein families, with N. nigricollis also containing four uncharacterized proteins. Conclusions: This study highlights venom compositional differences, advancing snake venom biology and informing targeted antivenom development.
Keywords: antivenom development; envenomation; iTRAQ; mass spectrometry; proteomics; snake venom.
Conflict of interest statement
The authors report no declarations of interest.
Figures





Similar articles
-
Preclinical evaluation of the neutralising efficacy of three antivenoms against the venoms of the recently taxonomically partitioned E. ocellatus and E. romani.PLoS Negl Trop Dis. 2025 Aug 4;19(8):e0013371. doi: 10.1371/journal.pntd.0013371. eCollection 2025 Aug. PLoS Negl Trop Dis. 2025. PMID: 40758764 Free PMC article.
-
Venom proteomic analysis of medically important Nigerian viper Echis ocellatus and Bitis arietans snake species.Biochem Biophys Rep. 2021 Nov 2;28:101164. doi: 10.1016/j.bbrep.2021.101164. eCollection 2021 Dec. Biochem Biophys Rep. 2021. PMID: 34765747 Free PMC article.
-
Deciphering toxico-proteomics of Asiatic medically significant venomous snake species: A systematic review and interactive data dashboard.Toxicon. 2024 Nov 6;250:108120. doi: 10.1016/j.toxicon.2024.108120. Epub 2024 Oct 10. Toxicon. 2024. PMID: 39393539
-
Toxicological analyses of the venoms of Nigerian vipers Echis ocellatus and Bitis arietans.Trop Med Health. 2024 Jan 29;52(1):15. doi: 10.1186/s41182-024-00581-9. Trop Med Health. 2024. PMID: 38282015 Free PMC article.
-
Systematic review and meta-analysis on the efficacy of Indian polyvalent antivenom against the Indian snakes of clinical significance.Arch Toxicol. 2024 Feb;98(2):375-393. doi: 10.1007/s00204-023-03643-9. Epub 2023 Dec 28. Arch Toxicol. 2024. PMID: 38153416
References
-
- Fernández C E.A., Youssef P. Snakebites in the Americas: A Neglected Problem in Public Health. Curr. Trop. Med. Rep. 2024;11:19–27. doi: 10.1007/s40475-023-00309-5. - DOI
-
- World Health Organization Target Product Profiles for Animal Plasma-Derived Antivenoms: Antivenoms for Treatment of Snakebite Envenoming in Sub-Saharan Africa. apps.who.int. 2023. [(accessed on 11 November 2024)]. Available online: https://apps.who.int/iris/bitstream/handle/10665/369786/9789240074569-en....
-
- WHO Guidelines for the Prevention and Clinical Management of Snakebite in Africa. [(accessed on 20 December 2024)]. Available online: https://www.who.int/publications/i/item/9789290231684.
Grants and funding
LinkOut - more resources
Full Text Sources

