Proteomic Profiling Reveals Novel Molecular Insights into Dysregulated Proteins in Established Cases of Rheumatoid Arthritis
- PMID: 40700276
- PMCID: PMC12286002
- DOI: 10.3390/proteomes13030032
Proteomic Profiling Reveals Novel Molecular Insights into Dysregulated Proteins in Established Cases of Rheumatoid Arthritis
Abstract
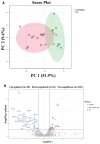
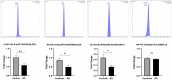
Background: Rheumatoid arthritis (RA) is a chronic autoimmune disorder that predominantly affects synovial joints, leading to inflammation, pain, and progressive joint damage. Despite therapeutic advancements, the molecular basis of established RA remains poorly defined. Methods: In this study, we conducted an untargeted plasma proteomic analysis using two-dimensional differential gel electrophoresis (2D-DIGE) and matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF-MS) in samples from RA patients and healthy controls in the discovery phase. Results: Significantly (ANOVA, p ≤ 0.05, fold change > 1.5) differentially abundant proteins (DAPs) were identified. Notably, upregulated proteins included mitochondrial dicarboxylate carrier, hemopexin, and 28S ribosomal protein S18c, while CCDC124, osteocalcin, apolipoproteins A-I and A-IV, and haptoglobin were downregulated. Receiver operating characteristic (ROC) analysis identified CCDC124, osteocalcin, and metallothionein-2 with high diagnostic potential (AUC = 0.98). Proteins with the highest selected frequency were quantitatively verified by multiple reaction monitoring (MRM) analysis in the validation cohort. Bioinformatic analysis using Ingenuity Pathway Analysis (IPA) revealed the underlying molecular pathways and key interaction networks involved STAT1, TNF, and CD40. These central nodes were associated with immune regulation, cell-to-cell signaling, and hematological system development. Conclusions: Our combined proteomic and bioinformatic approaches underscore the involvement of dysregulated immune pathways in RA pathogenesis and highlight potential diagnostic biomarkers. The utility of these markers needs to be evaluated in further studies and in a larger cohort of patients.
Keywords: 2D DIGE; MALDI-TOF; biomarker; metallothionein-2; osteocalcin; plasma proteomics; rheumatoid arthritis.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures







Similar articles
-
Impact of Tobacco Consumption on Salivary Protein Profiles: A MALDI-TOF Mass Spectrometry Study.Asian Pac J Cancer Prev. 2025 May 1;26(5):1483-1489. doi: 10.31557/APJCP.2025.26.5.1483. Asian Pac J Cancer Prev. 2025. PMID: 40439360 Free PMC article.
-
Shared molecular biomarkers and therapeutic targets in rheumatoid arthritis and osteoarthritis: Focus on EIF3B, KHSRP, NCL, PDCD1LG2, and SLC25A37.Cytokine. 2025 Sep;193:156988. doi: 10.1016/j.cyto.2025.156988. Epub 2025 Jun 30. Cytokine. 2025. PMID: 40592131
-
DS-Modified Paeoniflorin pH-Responsive Lipid-Polymer Hybrid Nanoparticles for Targeted Macrophage Polarization in a Rat Model of Rheumatoid Arthritis.Int J Nanomedicine. 2025 Jul 12;20:8967-8992. doi: 10.2147/IJN.S516434. eCollection 2025. Int J Nanomedicine. 2025. PMID: 40671689 Free PMC article.
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2021 Apr 19;4(4):CD011535. doi: 10.1002/14651858.CD011535.pub4. Cochrane Database Syst Rev. 2021. Update in: Cochrane Database Syst Rev. 2022 May 23;5:CD011535. doi: 10.1002/14651858.CD011535.pub5. PMID: 33871055 Free PMC article. Updated.
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2020 Jan 9;1(1):CD011535. doi: 10.1002/14651858.CD011535.pub3. Cochrane Database Syst Rev. 2020. Update in: Cochrane Database Syst Rev. 2021 Apr 19;4:CD011535. doi: 10.1002/14651858.CD011535.pub4. PMID: 31917873 Free PMC article. Updated.
References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

