Golgin Protein MoCoy1 Mediates Retrograde Trafficking From the Golgi to the ER, Regulating Fungal Development and Pathogenicity in Magnaporthe oryzae
- PMID: 40700342
- PMCID: PMC12285743
- DOI: 10.1111/mpp.70130
Golgin Protein MoCoy1 Mediates Retrograde Trafficking From the Golgi to the ER, Regulating Fungal Development and Pathogenicity in Magnaporthe oryzae
Abstract
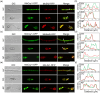
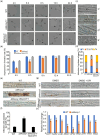
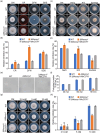
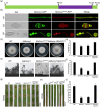
The Golgi apparatus is a vital organelle involved in protein sorting and trafficking. Golgins, a family of coiled-coil proteins, play an important role in maintaining the structure and function of the Golgi apparatus in eukaryotes. However, the function of golgins in the plant-pathogenic fungus Magnaporthe oryzae remains uncharacterised. Here, we systematically investigated the biological role of the golgin protein MoCoy1 in M. oryzae. Our results show that MoCoy1 is primarily localised to the Golgi apparatus. MoCOY1 deletion led to defects in vegetative growth, conidiation and pathogenicity of M. oryzae. In addition, MoCoy1 affected the secretion of the cytoplasmic effector proteins. Furthermore, MoCoy1 interacted with retrograde Golgi-related components and affected the retrograde transport from the Golgi to the endoplasmic reticulum (ER). Overall, our findings suggest that the golgin protein MoCoy1 mediates ER-Golgi retrograde trafficking, thereby regulating the development and pathogenicity of M. oryzae.
Keywords: Magnaporthe oryzae; Golgins; MoCoy1; pathogenicity; retrograde trafficking.
© 2025 The Author(s). Molecular Plant Pathology published by British Society for Plant Pathology and John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures




References
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources

