Prophage-encoded methyltransferase drives adaptation of community-acquired methicillin-resistant Staphylococcus aureus
- PMID: 40700354
- PMCID: PMC12435837
- DOI: 10.1172/JCI177872
Prophage-encoded methyltransferase drives adaptation of community-acquired methicillin-resistant Staphylococcus aureus
Abstract
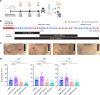
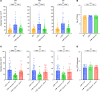
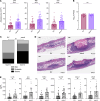
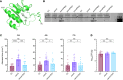
We recently described the evolution of a community-acquired methicillin-resistant Staphylococcus aureus (CA-MRSA) USA300 variant responsible for an outbreak of skin and soft tissue infections. Acquisition of a mosaic version of the Φ11 prophage (mΦ11) that increases skin abscess size was an early step in CA-MRSA adaptation that primed the successful spread of the clone. The present study shows how prophage mΦ11 exerts its effect on virulence for skin infection without encoding known toxin or fitness genes. Abscess size and skin inflammation were associated with DNA methylase activity of an mΦ11-encoded adenine methyltransferase (designated pamA). pamA increased expression of fibronectin-binding protein A (fnbA; FnBPA), and inactivation of fnbA eliminated the effect of pamA on abscess virulence without affecting strains lacking pamA. Thus, fnbA is a pamA-specific virulence factor. Mechanistically, pamA was shown to promote biofilm formation in vivo in skin abscesses, a phenotype linked to FnBPA's role in biofilm formation. Collectively, these data reveal a critical mechanism - epigenetic regulation of staphylococcal gene expression - by which phage can regulate virulence to drive adaptive leaps by S. aureus.
Keywords: Bacterial infections; Epigenetics; Infectious disease; Microbiology; Molecular biology.
Figures
Update of
-
Prophage-encoded methyltransferase drives adaptation of community-acquired methicillin-resistant Staphylococcus aureus.bioRxiv [Preprint]. 2024 Apr 17:2024.04.17.589803. doi: 10.1101/2024.04.17.589803. bioRxiv. 2024. Update in: J Clin Invest. 2025 Jul 22;135(18):e177872. doi: 10.1172/JCI177872. PMID: 38659881 Free PMC article. Updated. Preprint.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

