A nociceptive amygdala-striatal pathway modulating affective-motivational pain
- PMID: 40700496
- PMCID: PMC12285721
- DOI: 10.1126/sciadv.ado2837
A nociceptive amygdala-striatal pathway modulating affective-motivational pain
Abstract
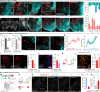
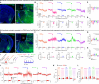
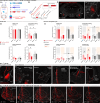
The basolateral amygdala (BLA) assigns valence to sensory stimuli, with a dedicated nociceptive ensemble encoding the negative valence of pain. However, the effects of chronic pain on the transcriptomic signatures and projection architecture of this BLA nociceptive ensemble are not well understood. Here, we show that optogenetic inhibition of the nociceptive BLA ensemble reduces affective-motivational behaviors in chronic neuropathic pain. Single-nucleus RNA sequencing revealed peripheral injury-induced changes in genetic pathways involved in axonal and presynaptic organization in nociceptive BLA neurons. Next, we identified a previously uncharacterized nociceptive hotspot in the nucleus accumbens shell that is innervated by BLA nociceptive neurons. Axonal calcium imaging of BLA projections to the accumbens and chemogenetic inhibition of this pathway revealed pain-related transmission from the amygdala to the medial nucleus accumbens, facilitating both acute and chronic pain affective-motivational behaviors. Together, this work defines a critical nociceptive amygdala-striatal circuit underlying pain unpleasantness across pain states.
Figures



Update of
-
A nociceptive amygdala-striatal pathway for chronic pain aversion.bioRxiv [Preprint]. 2024 Feb 13:2024.02.12.579947. doi: 10.1101/2024.02.12.579947. bioRxiv. 2024. Update in: Sci Adv. 2025 Jul 25;11(30):eado2837. doi: 10.1126/sciadv.ado2837. PMID: 38405972 Free PMC article. Updated. Preprint.
References
-
- Kuner R., Kuner T., Cellular circuits in the brain and their modulation in acute and chronic pain. Physiol. Rev. 101, 213–258 (2021). - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources

