N4BP1 is a nucleocytoplasmic shuttling protein and recognizes aggregates of the ubiquitin-like protein NEDD8 to protect cells under heat shock
- PMID: 40701250
- PMCID: PMC12365338
- DOI: 10.1016/j.jbc.2025.110511
N4BP1 is a nucleocytoplasmic shuttling protein and recognizes aggregates of the ubiquitin-like protein NEDD8 to protect cells under heat shock
Abstract
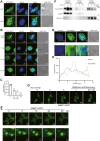
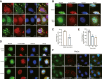
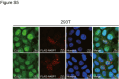
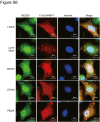
N4BP1 (NEDD4-binding partner 1) is a key checkpoint for proper inflammatory responses; however, its cellular localization, biologic nature, and functions in other cellular processes remain largely unknown. In this study, we demonstrate that N4BP1 is a nucleocytoplasmic shuttling protein. Treatment with leptomycin B induces nuclear accumulation of N4BP1, and the region responsible for its nuclear distribution maps to amino acids 151 to 338. Further analysis identified a nuclear localization signal (NLS) spanning residues 279 to 299. Deletion or mutation of this NLS abolishes N4BP1 nuclear import, while fusing the NLS to GFP is sufficient to drive GFP into the nucleus. Notably, we found that N4BP1 forms protein aggregates in both the cytoplasm and nucleus. These aggregates lack ubiquitin-modified proteins but instead colocalize with NEDD8-modified proteins. Consistently, N4BP1 aggregates contain cullin-1 and cullin-2. The CoCUN domain is essential for recognizing neddylated proteins and mediating N4BP1 aggregate formation. N4BP1 aggregates exhibit liquid-liquid phase separation, as evidenced by their sensitivity to 1,6-hexanediol (an liquid-liquid phase separation inhibitor). Smaller N4BP1 aggregates can fuse into larger one and reassemble after 1,6-hexanediol-induced disruption. Furthermore, heat shock promotes N4BP1 aggregation, which confers cellular protection under stress conditions. Taken together, our findings reveal that N4BP1 is a nucleocytoplasmic shuttling protein. N4BP1 forms protein aggregates that contain neddylated proteins such as cullin-1 and cullin-2. This study uncovers the previously unrecognized role of N4BP1 in organizing neddylated protein aggregates and highlights its functional significance in stress adaption.
Keywords: N4BP1; NEDD8; aggregates; heat shock; nucleocytoplasmic shuttling.
Copyright © 2025 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare that they have no conflict of interest with the contents of this article.
Figures










Similar articles
-
Enteroviral 3C protease cleaves N4BP1 to impair the host inflammatory response.J Virol. 2025 Jan 31;99(1):e0175824. doi: 10.1128/jvi.01758-24. Epub 2024 Dec 10. J Virol. 2025. PMID: 39655957 Free PMC article.
-
Prescription of Controlled Substances: Benefits and Risks.2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 30726003 Free Books & Documents.
-
Electrophoresis.2025 Jul 14. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2025 Jul 14. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 36251838 Free Books & Documents.
-
Management of urinary stones by experts in stone disease (ESD 2025).Arch Ital Urol Androl. 2025 Jun 30;97(2):14085. doi: 10.4081/aiua.2025.14085. Epub 2025 Jun 30. Arch Ital Urol Androl. 2025. PMID: 40583613 Review.
-
Immunogenicity and seroefficacy of pneumococcal conjugate vaccines: a systematic review and network meta-analysis.Health Technol Assess. 2024 Jul;28(34):1-109. doi: 10.3310/YWHA3079. Health Technol Assess. 2024. PMID: 39046101 Free PMC article.
References
-
- Murillas R., Simms K.S., Hatakeyama S., Weissman A.M., Kuehn M.R. Identification of developmentally expressed proteins that functionally interact with Nedd4 ubiquitin ligase. J. Biol. Chem. 2002;277:2897–2907. - PubMed
-
- Gitlin A.D., Heger K., Schubert A.F., Reja R., Yan D., Pham V.C., et al. Integration of innate immune signalling by caspase-8 cleavage of N4BP1. Nature. 2020;587:275–280. - PubMed
-
- Wu C., Guo X., Zheng W., Sun R., Chen L., Shen Y., et al. N4BP1 regulates keratinocytes development and plays protective role in burn- and adhesive-related skin injury via MMP9. Cell Signal. 2023;110 - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

