Multiple Autopolyploid Arabidopsis lyrata Populations Stabilized by Long-Range Adaptive Introgression Across Eurasia
- PMID: 40701836
- PMCID: PMC12342985
- DOI: 10.1093/molbev/msaf153
Multiple Autopolyploid Arabidopsis lyrata Populations Stabilized by Long-Range Adaptive Introgression Across Eurasia
Abstract
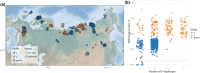
Abundance of established polyploid lineages varies across lineages, evolutionary time, and geography, suggesting both genetics and environment play a role in polyploid persistence. We show Arabidopsis lyrata is the most polyploid-rich species complex in the Arabidopsis genus, with multiple origins of autotetraploidy. This is revealed by genomic data from over 400 A. lyrata samples across Eurasia. We found over 30 previously undescribed autotetraploid populations in Siberia with a minimum of two separate origins, independent of those previously reported in Central Europe. The establishment of Siberian tetraploids is mediated by meiotic adaptation at the same genes as in European tetraploid A. lyrata and Arabidopsis arenosa, despite their genomic divergence and geographical separation. Haplotype analysis based on synthetic long-read assemblies supports the long-range introgression of adaptive alleles from the tetraploid interspecific pool of European A. lyrata and A. arenosa to tetraploid Siberian A. lyrata. Once adaptations to polyploidy emerge, they promote the establishment of new polyploid lineages through adaptive inter- and intraspecific introgression.
Keywords: Arabidopsis lyrata; adaptation; introgression; polyploid.
© The Author(s) 2025. Published by Oxford University Press on behalf of Society for Molecular Biology and Evolution.
Figures


Similar articles
-
Interspecific and interploidal gene flow in Central European Arabidopsis (Brassicaceae).BMC Evol Biol. 2011 Nov 29;11:346. doi: 10.1186/1471-2148-11-346. BMC Evol Biol. 2011. PMID: 22126410 Free PMC article.
-
Prescription of Controlled Substances: Benefits and Risks.2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 30726003 Free Books & Documents.
-
Indian Himalayan Arabidopsis thaliana population reflects deep history and might be the source of species expansion in the eastern edge.BMC Plant Biol. 2025 Jul 29;25(1):980. doi: 10.1186/s12870-025-06944-6. BMC Plant Biol. 2025. PMID: 40731258 Free PMC article.
-
Signs and symptoms to determine if a patient presenting in primary care or hospital outpatient settings has COVID-19.Cochrane Database Syst Rev. 2022 May 20;5(5):CD013665. doi: 10.1002/14651858.CD013665.pub3. Cochrane Database Syst Rev. 2022. PMID: 35593186 Free PMC article.
-
Falls prevention interventions for community-dwelling older adults: systematic review and meta-analysis of benefits, harms, and patient values and preferences.Syst Rev. 2024 Nov 26;13(1):289. doi: 10.1186/s13643-024-02681-3. Syst Rev. 2024. PMID: 39593159 Free PMC article.
References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

