UBE4B promotes gastric cancer proliferation and metastasis by mediating FAT4 ubiquitination and degradation
- PMID: 40701960
- PMCID: PMC12287395
- DOI: 10.1038/s41419-025-07794-8
UBE4B promotes gastric cancer proliferation and metastasis by mediating FAT4 ubiquitination and degradation
Abstract
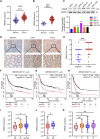
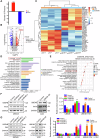
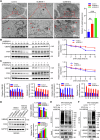
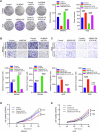
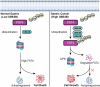
The ubiquitin‒proteasome system (UPS), an intracellular protein degradation pathway, plays an important role in regulating tumorigenesis and development. Ubiquitination factor E4B (UBE4B/UFD2) has been shown to be associated with the development of several cancers. The aim of this study was to reveal the functional significance of UBE4B in gastric cancer (GC) development and its important mechanism. Bioinformatics analysis, immunohistochemistry (IHC), western blotting, and real-time PCR were performed to detect UBE4B expression in human GC samples and GC cell lines and a mouse xenograft tumour model was established. Our investigation revealed that UBE4B is highly expressed in GC and promotes the proliferation, migration and invasion of GC cells. The quantitative Tandem Mass Tag (TMT) analysis revealed that FAT oncogenic homologue 4 (FAT4) is a downstream gene of UBE4B. Western blot experiments and transmission electron microscopy (TEM) results for biological samples revealed that UBE4B inhibits autophagy in GC cells and directly binds to and degrades FAT4 through ubiquitination. These results suggest that UBE4B can inhibit autophagy and promote GC progression by mediating FAT4 ubiquitination and degradation, and our findings provide a new potential therapeutic target for GC management.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests. Ethical approval: Inclusion of human participants, and use of human data and human tissue in this study were approved by the Ethics Committee of the First Affiliated Hospital of Nanchang University. The use of animals in this study was approved by the animal research committee in the First Affiliated Hospital of Nanchang University.
Figures




References
-
- Bray F, Laversanne M, Sung H, Ferlay J, Siegel RL, Soerjomataram I, et al. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2024;74:229–63. - PubMed
-
- Herrero R, Parsonnet J, Greenberg ER. Prevention of gastric cancer. Jama. 2014;312:1197–8. - PubMed
-
- Wang Y, Le WD. Autophagy and ubiquitin-proteasome system. Adv Exp Med Biol. 2019;1206:527–50. - PubMed
MeSH terms
Substances
Grants and funding
- 81760524/National Natural Science Foundation of China (National Science Foundation of China)
- 82260599/National Natural Science Foundation of China (National Science Foundation of China)
- 81760524/National Natural Science Foundation of China (National Science Foundation of China)
- 82260599/National Natural Science Foundation of China (National Science Foundation of China)
- 81760524/National Natural Science Foundation of China (National Science Foundation of China)
- 82260599/National Natural Science Foundation of China (National Science Foundation of China)
- 81760524/National Natural Science Foundation of China (National Science Foundation of China)
- 82260599/National Natural Science Foundation of China (National Science Foundation of China)
- 81760524/National Natural Science Foundation of China (National Science Foundation of China)
- 82260599/National Natural Science Foundation of China (National Science Foundation of China)
- 81760524/National Natural Science Foundation of China (National Science Foundation of China)
- 82260599/National Natural Science Foundation of China (National Science Foundation of China)
- 81760524/National Natural Science Foundation of China (National Science Foundation of China)
- 82260599/National Natural Science Foundation of China (National Science Foundation of China)
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

