Proteasome inhibition overcomes resistance to targeted therapies in B-cell malignancy models and in an index patient
- PMID: 40701968
- PMCID: PMC12287370
- DOI: 10.1038/s41419-025-07884-7
Proteasome inhibition overcomes resistance to targeted therapies in B-cell malignancy models and in an index patient
Abstract
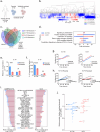
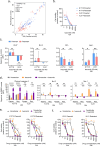
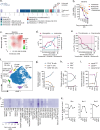
Treatment of B-cell malignancies with the PI3K inhibitor (PI3Ki) idelalisib often results in high toxicity and resistance, with limited treatment alternatives for relapsed/refractory patients since idelalisib is recommended as a later or last line therapy. To investigate resistance mechanisms and identify alternative treatments, we studied functional phenotypes of idelalisib-resistant B-cell malignancy models. The idelalisib-resistant KARPAS1718 model remained sensitive to Bcl-2 inhibitors (Bcl-2i), whereas the resistant VL51 model showed reduced sensitivity compared to parental cells. Sensitivity correlated with phosphorylation and expression of the Bcl-2 family members Bcl-2 and Bim. Target addiction scoring revealed high dependence on the proteasome, and proteasome inhibitors (PI) were effective across models and in primary chronic lymphocytic leukemia (CLL) cells, independently of their PI3Ki- or Bcl-2i-sensitivities. PI treatment consistently upregulated Bim and Mcl-1, while Bcl-2 increased in KARPAS1718 and CLL cells. Bcl-2i plus PI combinations led to an additive effect in these models. A multi-refractory CLL patient in the IMPRESS-Norway trial (NCT04817956) treated with Bcl-2i plus PI showed initial clinical improvement but relapsed within four months. Treatment induced Bim and Mcl-1 upregulation and reduced cytotoxic CD8+ T-cell and CD56dim NK-cell populations. Our findings suggest that PIs may overcome resistance to targeted therapies, and warrant further studies to optimize clinical responses.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: ÅH has received research support from AstraZeneca, BMS, EliLilly, GSK, Incyte, Novartis, Roche, Ultimovacs, the Norwegian Cancer Society, The regional health authorities in Norway. Have given talks and advise to pharma companies, and all honoraria are going to the hospital (Abbvie, AstraZeneca, BMS, EliLilly, Janssen, Medicover, Merck, Novartis, Pfizer, Roche, Sanofi, Takeda). ARM has had a consulting or advisory role for AstraZeneca, TG Therapeutics, AbbVie/Genentech, Pharmacyclics, Adaptive Biotechnologies, Johnson & Johnson, Acerta Pharma/AstraZeneca, Loxo/Lilly, Curio/Vaniam Group, Merck, Bristol Myers Squibb/Pfizer, PerView, DAVA Oncology, BMS, Genmab, AXIS Education, and PER; and has received research funding from Regeneron, TG Therapeutics, Sunesis Pharmaceuticals, Loxo, AbbVie/Genentech, Pharmacyclics, Adaptive Biotechnologies, Johnson & Johnson, Acerta Pharma/AstraZeneca, DTRM, Genmab, and Nurix. FB has received institutional research funds from ADC Therapeutics, Bayer AG, BeiGene, Floratek Pharma, Helsinn, HTG Molecular Diagnostics, Ideogen AG, Idorsia Pharmaceuticals Ltd., Immagene, ImmunoGen, Menarini Ricerche, Nordic Nanovector ASA, Oncternal Therapeutics, Spexis AG; consultancy fee from BIMINI Biotech, Floratek Pharma, Helsinn, Immagene, Menarini, Vrise Therapeutics; advisory board fees to institution from Novartis; expert statements provided to HTG Molecular Diagnostics; travel grants from Amgen, Astra Zeneca, iOnctura. GET has received research grants from Mundipharma and Alexion Pharmaceuticals, advisory board honoraria from Alexion Pharmaceuticals and Janssen-Cilag, and lecture honoraria from Novartis, Janssen-Cilag, Alexion Pharmaceuticals, and Mundipharma. SSS has received consulting fees from AstraZeneca, BeiGene and Janssen; and research support from BeiGene and TG Therapeutics. The other authors declare no competing financial interests.
Figures


Similar articles
-
BCL-2 dependence is a favorable predictive marker of response to therapy for chronic lymphocytic leukemia.Mol Cancer. 2025 Mar 3;24(1):62. doi: 10.1186/s12943-025-02260-7. Mol Cancer. 2025. PMID: 40025512 Free PMC article.
-
Indirect Treatment Comparisons of Ibrutinib Versus Physician's Choice and Idelalisib Plus Ofatumumab in Patients With Previously Treated Chronic Lymphocytic Leukemia.Clin Ther. 2017 Jan;39(1):178-189.e5. doi: 10.1016/j.clinthera.2016.12.001. Epub 2017 Jan 3. Clin Ther. 2017. PMID: 28062113
-
Therapeutic advances in chronic lymphocytic leukemia: A focus on molecular pathogenesis, targeted therapies, and supportive care.Am J Health Syst Pharm. 2025 Aug 5;82(16):868-885. doi: 10.1093/ajhp/zxaf058. Am J Health Syst Pharm. 2025. PMID: 40071479 Review.
-
LP-118 is a novel B-cell lymphoma 2 / extra-large inhibitor that demonstrates efficacy in models of venetoclaxresistant chronic lymphocytic leukemia.Haematologica. 2025 Jan 1;110(1):78-91. doi: 10.3324/haematol.2023.284353. Haematologica. 2025. PMID: 39113656 Free PMC article.
-
Rituximab, ofatumumab and other monoclonal anti-CD20 antibodies for chronic lymphocytic leukaemia.Cochrane Database Syst Rev. 2012 Nov 14;11(11):CD008079. doi: 10.1002/14651858.CD008079.pub2. Cochrane Database Syst Rev. 2012. PMID: 23152253 Free PMC article.
References
-
- Richardson NC, Kasamon Y, Pazdur R, Gormley N. The saga of PI3K inhibitors in haematological malignancies: survival is the ultimate safety endpoint. Lancet Oncol. 2022;23:563–6. - PubMed
-
- Eichhorst B, Ghia P, Niemann CU, Kater AP, Gregor M, Hallek M, et al. ESMO Clinical Practice Guideline interim update on new targeted therapies in the first-line and at relapse of chronic lymphocytic leukaemia. Ann Oncol J Eur Soc Med Oncol. 2024;35:762–8. - PubMed
-
- Aronson JH, Skånland SS, Roeker LE, Thompson MC, Mato AR. Approach to a patient with “double refractory” chronic lymphocytic leukemia: “Double, double toil and trouble” (Shakespeare). Am J Hematol. 2022;97:S19–25. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

