Targeting SLC7A5 in lung squamous cell carcinoma: implications for cancer metabolism shift and boron neutron capture therapy resistance
- PMID: 40701982
- PMCID: PMC12287270
- DOI: 10.1038/s41389-025-00568-z
Targeting SLC7A5 in lung squamous cell carcinoma: implications for cancer metabolism shift and boron neutron capture therapy resistance
Abstract
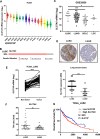
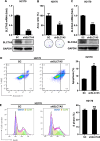
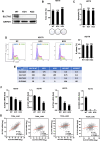
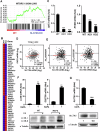
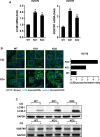
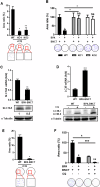
Squamous cell carcinoma (SCC) poses a significant global health challenge due to the lack of effective treatments. Boron neutron capture therapy (BNCT), a targeted particle therapy, has shown promising results in various cancers. SLC7A5, a transporter of essential amino acids and boronophenylalanine (BPA) used in BNCT, emerges as a potential therapeutic target. However, its expression across different histological subtypes and the role of SLC7A5 inhibition in developing drug resistance to BPA-BNCT remain poorly understood. Our study reveals elevated SLC7A5 expression in most SCCs, particularly in lung squamous cell carcinoma (LUSC), where it is significantly higher compared to other lung cancer subtypes. Increased SLC7A5 expression and a higher tumor-to-normal (T/N) ratio in LUSC are associated with poor patient prognosis. SLC7A5 knockdown in LUSC cells reduces colony formation and induces apoptosis. RNA-seq analysis of SLC7A5 knockout LUSC cells shows downregulated mTORC1 signaling, reduced expression of other amino acid transporters, and upregulated autophagy genes, indicating a potential cancer metabolic shift. Furthermore, SLC7A5 knockout LUSC cells demonstrate resistance to BPA-BNCT but sensitivity to the autophagy inhibitor chloroquine. Post-BPA-BNCT treatment, surviving wild-type LUSC cells exhibit reduced SLC7A5 levels and increased sensitivity to chloroquine, highlighting a vulnerability in BPA-BNCT-resistant cells. Our findings elucidate the interplay between SLC7A5, mTOR signaling, and autophagy pathways, providing insights into potential strategies to overcome drug resistance in BPA-BNCT therapy.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures
References
-
- Kanai Y, Segawa H, Miyamoto K, Uchino H, Takeda E, Endou H. Expression cloning and characterization of a transporter for large neutral amino acids activated by the heavy chain of 4F2 antigen (CD98). J Biol Chem. 1998;273:23629–32. - PubMed
-
- Kanai Y. Amino acid transporter LAT1 (SLC7A5) as a molecular target for cancer diagnosis and therapeutics. Pharm Ther. 2022;230:107964. - PubMed
-
- Kim CS, Cho SH, Chun HS, Lee SY, Endou H, Kanai Y, et al. BCH, an inhibitor of system L amino acid transporters, induces apoptosis in cancer cells. Biol Pharm Bull. 2008;31:1096–100. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

