The neural basis of species-specific defensive behaviour in Peromyscus mice
- PMID: 40702175
- PMCID: PMC12422964
- DOI: 10.1038/s41586-025-09241-2
The neural basis of species-specific defensive behaviour in Peromyscus mice
Abstract
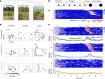
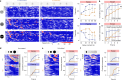
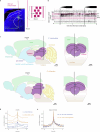
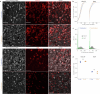
Evading imminent threat from predators is critical for animal survival. Effective defensive strategies can vary, even between closely related species. However, the neural basis of such species-specific behaviours remains poorly understood1-4. Here we find that two sister species of deer mice (genus Peromyscus)5 show different responses to the same looming stimulus: Peromyscus maniculatus, which occupies densely vegetated habitats, predominantly escapes, whereas the open field specialist, Peromyscus polionotus, briefly freezes. This difference arises from species-specific escape thresholds, is largely context-independent, and can be triggered by both visual and auditory threat stimuli. Using immunohistochemistry and electrophysiological recordings, we find that although visual threat activates the superior colliculus in both species, the role of the dorsal periaqueductal grey (dPAG) in driving behaviour differs. Whereas dPAG activity scales with running speed in P. maniculatus, neural activity in the dPAG of P. polionotus correlates poorly with movement, including during visually triggered escape. Moreover, optogenetic activation of dPAG neurons elicits acceleration in P. maniculatus but not in P. polionotus, and their chemogenetic inhibition during a looming stimulus delays escape onset in P. maniculatus to match that of P. polionotus. Together, we trace species-specific escape thresholds to a central circuit node, downstream of peripheral sensory neurons, localizing an ecologically relevant behavioural difference to a specific region of the mammalian brain.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures











Update of
-
The neural basis of defensive behaviour evolution in Peromyscus mice.bioRxiv [Preprint]. 2023 Jul 5:2023.07.04.547734. doi: 10.1101/2023.07.04.547734. bioRxiv. 2023. Update in: Nature. 2025 Sep;645(8080):439-447. doi: 10.1038/s41586-025-09241-2. PMID: 37461474 Free PMC article. Updated. Preprint.
References
-
- Wada-Katsumata, A., Silverman, J. & Schal, C. Changes in taste neurons support the emergence of an adaptive behavior in cockroaches. Science340, 972–975 (2013). - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

