Deciphering phenylalanine-derived salicylic acid biosynthesis in plants
- PMID: 40702180
- PMCID: PMC12408371
- DOI: 10.1038/s41586-025-09280-9
Deciphering phenylalanine-derived salicylic acid biosynthesis in plants
Abstract
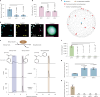
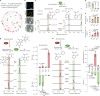
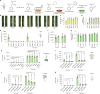
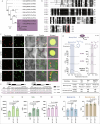
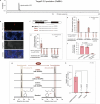
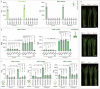
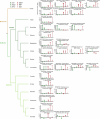
Salicylic acid (SA) is a ubiquitous plant hormone with a long history in human civilization1,2. Because of the central role of SA in orchestrating plant pathogen defence, understanding SA biosynthesis is fundamental to plant immunity research and crop improvement. Isochorismate-derived SA biosynthesis has been well defined in Arabidopsis. However, increasing evidence suggests a crucial function for phenylalanine-derived SA biosynthesis in many other plant species1. Here we reveal the phenylalanine-derived SA biosynthetic pathway in rice by identifying three dedicated enzymes - peroxisomal benzoyl-CoA:benzyl alcohol benzoyltransferase (BEBT), the endoplasmic reticulum-associated cytochrome P450 enzyme benzylbenzoate hydroxylase (BBH), and cytosolic benzylsalicylate esterase (BSE) that sequentially convert benzoyl-CoA to benzylbenzoate, benzylsalicylate and SA. The pathogen-induced gene expression pattern and SA biosynthetic functions of this triple-enzyme module are conserved in diverse plants. This work fills a major knowledge gap in the biosynthesis of a key plant defence hormone, establishing a foundation for new strategies to create disease-resistant crops.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures







References
-
- Ullah, C., Chen, Y.-H., Ortega, M. A. & Tsai, C.-J. The diversity of salicylic acid biosynthesis and defense signaling in plants: knowledge gaps and future opportunities. Curr. Opin. Plant Biol.72, 102349 (2023). - PubMed
-
- Desborough, M. J. R. & Keeling, D. M. The aspirin story—from willow to wonder drug. Br. J. Haematol.177, 674–683 (2017). - PubMed
-
- Zou, Z. et al. Biochemical pathways of salicylic acid derived from l-phenylalanine in plants with different basal SA levels. J. Agric. Food Chem.72, 2898–2910 (2024). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

