A gut sense for a microbial pattern regulates feeding
- PMID: 40702192
- PMCID: PMC12443592
- DOI: 10.1038/s41586-025-09301-7
A gut sense for a microbial pattern regulates feeding
Abstract
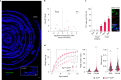
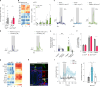
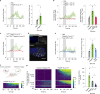
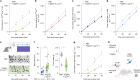
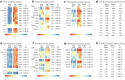
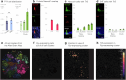
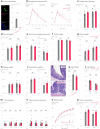
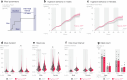
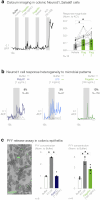
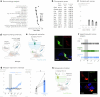
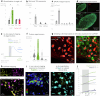
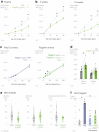
To coexist with its resident microorganisms, the host must have a sense to adjust its behaviour in response to them. In the intestine, a sense for nutrients transduced to the brain through neuroepithelial circuits guides appetitive choices1-5. However, a sense that allows the host to respond in real time to stimuli arising from resident gut microorganisms remains to be uncovered. Here we show that in the mouse colon, the ubiquitous microbial pattern flagellin-a unifying feature across phyla6-stimulates Toll-like receptor 5 (TLR5) in peptide YY (PYY)-labelled colonic neuropod cells. This stimulation leads to PYY release onto NPY2R vagal nodose neurons to regulate feeding. Mice lacking TLR5 in these cells eat more and gain more weight than controls. We found that flagellin does not act on the nerve directly. Instead, flagellin stimulates neuropod cells from the colonic lumen to reduce feeding through a gut-brain sensory neural circuit. Moreover, flagellin reduces feeding independent of immune responses, metabolic changes or the presence of gut microbiota. This sense enables the host to adjust its behaviour in response to a molecular pattern from its resident microorganisms. We call this sense at the interface of the biota and the brain the neurobiotic sense7.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: D.V.B. and M.M.K. are founders and board directors of the Gastronauts Foundation, Inc, a 501(c)3 non-profit company. Some of the findings have been filed by D.V.B. as a provisional patent application. All other authors declare no competing interests.
Figures

References
MeSH terms
Substances
Grants and funding
- DP2 MH122402/MH/NIMH NIH HHS/United States
- F32 DK127757/DK/NIDDK NIH HHS/United States
- F30 DK122712/DK/NIDDK NIH HHS/United States
- R21 AT010818/AT/NCCIH NIH HHS/United States
- F30 DK127650/DK/NIDDK NIH HHS/United States
- K01 DK138286/DK/NIDDK NIH HHS/United States
- F30 DK136229/DK/NIDDK NIH HHS/United States
- R01 DK131112/DK/NIDDK NIH HHS/United States
- R03 DK114500/DK/NIDDK NIH HHS/United States
- K01 DK131403/DK/NIDDK NIH HHS/United States
- F32 DK139628/DK/NIDDK NIH HHS/United States
- R01 DK132070/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

