Early methionine availability attenuates T cell exhaustion
- PMID: 40702340
- PMCID: PMC12307228
- DOI: 10.1038/s41590-025-02223-6
Early methionine availability attenuates T cell exhaustion
Abstract
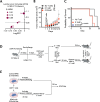
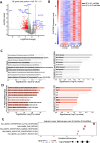
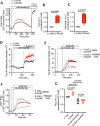
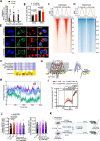
T cell receptor (TCR) activation is regulated in many ways, including niche-specific nutrient availability. Here we investigated how methionine (Met) availability and TCR signaling interplay during the earliest events of T cell activation affect subsequent cell fate. Limiting Met during the initial 30 min of TCR engagement increased Ca2+ influx, NFAT1 (encoded by Nfatc2) activation and promoter occupancy, leading to T cell exhaustion. We identified changes in the protein arginine methylome during initial TCR engagement and identified an arginine methylation of the Ca2+-activated potassium transporter KCa3.1, which regulates Ca2+-mediated NFAT1 signaling for optimal activation. Ablation of KCa3.1 arginine methylation increased NFAT1 nuclear localization, rendering T cells dysfunctional in mouse tumor and infection models. Furthermore, acute, early Met supplementation reduced nuclear NFAT1 in tumor-infiltrating T cells and augmented antitumor activity. These findings identify a metabolic event early after T cell activation that affects cell fate.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: E.S.G. consults for Anviron and G.K. has patent applications in the fields of cell or gene therapy for cancer and has received honoraria from Cell Signaling and Kineticos within the last 2 years. During the course of this research, D.R.G. consulted for Sonata Therapeutics, Ventus Therapeutics, Mycos and ASHA Therapeutics, and received honoraria, travel and/or research support from Horizon/Amgen, Lilly and Boeringher-Ingleheim. The other authors declare no competing interests.
Figures











Update of
-
An early, novel arginine methylation of KCa3.1 attenuates subsequent T cell exhaustion.bioRxiv [Preprint]. 2024 Nov 20:2024.05.09.593421. doi: 10.1101/2024.05.09.593421. bioRxiv. 2024. Update in: Nat Immunol. 2025 Aug;26(8):1384-1396. doi: 10.1038/s41590-025-02223-6. PMID: 38798680 Free PMC article. Updated. Preprint.
References
MeSH terms
Substances
Grants and funding
- R01 AI123322/AI/NIAID NIH HHS/United States
- K99 CA256262/CA/NCI NIH HHS/United States
- AI123322/U.S. Department of Health & Human Services | NIH | National Institute of Allergy and Infectious Diseases (NIAID)
- K99CA256262/U.S. Department of Health & Human Services | NIH | National Cancer Institute (NCI)
- CA231620/U.S. Department of Health & Human Services | NIH | National Cancer Institute (NCI)
LinkOut - more resources
Full Text Sources
Miscellaneous

