Unraveling resistance mechanisms to the novel nucleoside analog RX-3117 in lung cancer: insights into DNA repair, cell cycle dysregulation and targeting PKMYT1 for improved therapy
- PMID: 40702552
- PMCID: PMC12288264
- DOI: 10.1186/s13046-025-03470-z
Unraveling resistance mechanisms to the novel nucleoside analog RX-3117 in lung cancer: insights into DNA repair, cell cycle dysregulation and targeting PKMYT1 for improved therapy
Abstract
Background: Nucleoside analogues are crucial in treating non-small cell lung cancer (NSCLC), but resistance hampers patient outcomes. The cytidine analogue RX-3117 shows promise in gemcitabine-resistant cancers, yet mechanisms underlying acquired resistance to this drug remain unexplored. This study includes a comprehensive investigation into RX-3117 resistance mechanisms by leveraging new preclinical models and cutting-edge genomic tools, including a CRISPR-Cas9 knockout screen and transcriptomics.
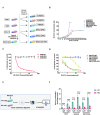
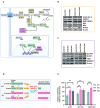
Methods: NSCLC cell lines A549 and SW1573 were exposed to stepwise increasing concentrations of RX-3117 to establish stable resistant subclones, confirmed by SRB and clonogenic assays. Intracellular RX-3117 nucleotide levels were measured via LC/MS-MS, prompting the evaluation and modulation of the expression of key metabolic enzymes by Western blot and siRNA. A CRISPR-Cas9 screen identified genes whose loss increased RX-3117 sensitivity, while RNA-sequencing with differential expression analyses revealed resistance-related pathways, further investigated through cell cycle distribution, knock-out, and ELISA assays.
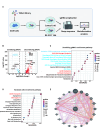
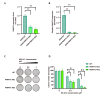
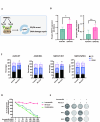
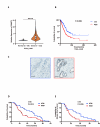
Results: Resistant clones exhibited decreased accumulation of RX-3117 nucleotides, which however, was not associated to reduced expression of activation enzymes (UCK2, UMPK, CMPK, NME1/NDPK, RR1 and RR2). Instead, increased expression was observed in certain DNA repair and deactivation enzymes (NT5C3) but pharmacological inhibition and silencing of the latter did not circumvent resistance. Remarkably, a comprehensive approach with CRISPR-Cas9 screen highlighted DNA-repair and cell cycle determinants as key sensitizing genes. XL-PCR and RNA-sequencing confirmed aberrations in DNA-repair and pathways involved in cell cycle regulation. Knock-out and pharmacological inhibition validated the role of PKMYT1, a protein kinase involved in G2/M transition and genomic stability. RX-3117-resistant A549 cells showed enhanced sensitivity to the PKMYT1 inhibitor lunresertib and its synergism with RX-3117, suggesting further studies, especially in patients with high PKMYT1 expression who have significantly shorter survival rates, as observed in public databases and validated in an internal cohort of NSCLC patients.
Conclusion: By integrating CRISPR-Cas9 with functional assays and transcriptomics, our study established a framework for decoding resistance mechanisms and highlights potential therapeutic strategies to enhance RX-3117 efficacy in NSCLC. We demonstrated for the first time that aberrant DNA repair and cell cycle dysregulation led resistance, identifying PKMYT1 as a promising target.
Keywords: Cell cycle distribution; Chemoresistance; DNA repair; Non-small cell lung cancer; Nucleoside analogs; PKMYT1.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: The study on patients’ specimens was approved by the local ethics committees (Ethics Committee of Istituto Clinico Humanitas (Rozzano, Milan, Italy)– Ref-No. 165/17, Comitato Etico di Area Vasta Nord Ovest (CEAVNO) Regione Toscana– Protocol No. 31677, University Hospital Antwerp (Antwerp, Belgium) ethics committee– Protocol No. B300201316249 and Onze Lieve Vrouw Hospital (Aalst, Belgium) ethics committee– Protocol No. B300201317801) and conducted in accordance with principles stated in the Declaration of Helsinki. Written informed consent was obtained from all patients. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures
References
-
- Elnaggar M, Giovannetti E, Peters GJ. Molecular targets of gemcitabine action: rationale for development of novel drugs and drug combinations. Curr Pharm Des. 2012;18(19):2811–29. - PubMed
-
- Trial.gov C. Dose-Finding and Safety Study for Oral Single-Agent to Treat Advanced Malignancies: ClinicalTrials.gov; [Available from: https://www.clinicaltrials.gov/study/NCT02030067?intr=RX-3117&rank=2
-
- Salgia N, Pal SK, Chung V, Tagawa ST, Picus J, Babiker HM et al. Activity of RX-3117, an oral antimetabolite nucleoside, in subjects with advanced urothelial cancer: preliminary results of a phase IIa study. J Clin Oncol. 2019;37(7).
-
- Babiker H, Schlegel PJ, Hicks LG, Bullock AJ, Burhani N, Mahadevan D, et al. A multicenter phase 1/2 study investigating the safety, pharmacokinetics, pharmacodynamics and efficacy of a small molecule antimetabolite, RX-3117, plus nab-paclitaxel in pancreatic adenocarcinoma. Invest New Drugs. 2022;40(1):81–90. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

