Exploring potential mechanisms of an African protective locus for Alzheimer's disease in APOEε4 carriers
- PMID: 40704449
- PMCID: PMC12287753
- DOI: 10.1002/alz.70500
Exploring potential mechanisms of an African protective locus for Alzheimer's disease in APOEε4 carriers
Abstract
Introduction: We recently described that the African-specific A allele of rs10423769, which lies in an area of segmental duplications, reduces Alzheimer's disease (AD) risk by ∼ 75% in apolipoprotein E (APOE) ε4 homozygotes.
Methods: Short and long-read sequencing were used to identify the haplotype harboring rs10423769_A and examine DNA methylation and structural variation (SV).
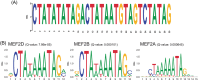
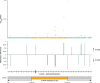
Results: A unique 21 kb haplotype is shared amongst all rs10423769_A carriers (r2 > 0.95) and is present in all African ancestry tested. We identified methylation differences between the non-protective and rs10423769_A haplotypes. An expanded variable number tandem repeat (VNTR) containing multiple MEF2-family transcription factor binding motifs in LD with the protective haplotype. Further, rs10423769 is an eQTL for ZNF222.
Discussion: The protective haplotype is unique, with haplotype changes in DNA methylation and SV that could contribute to the protective mechanism of rs10423769_A. The study generates hypothesis for future studies on this important protective mechanism for APOEε4 carriers.
Highlights: We investigated an African-specific haplotype harboring a protective locus for APOEε4 carriers that significantly reduces the risk for Alzheimer Disease. The protective locus lies in an area of segmental duplications 2MB from APOE. We found that the protective haplotype is unique within the segmental duplications. The haplotype is found in all African ancestry tested but not in other ancestries. Long read whole genome sequencing identified an expanded VNTR associated with the protective haplotype containing multiple MEF2 transcription factor binding motifs. Differences in DNA methylation were found between the protective and the reference haplotype.
Keywords: Alzheimer's disease; genetic ancestry; haplotype; long‐read sequencing; methylation; protective variant; structural variation.
© 2025 The Author(s). Alzheimer's & Dementia published by Wiley Periodicals LLC on behalf of Alzheimer's Association.
Conflict of interest statement
The authors declare no competing interests.
Figures





Update of
-
African origin haplotype protective for Alzheimer's disease in APOEε4 carriers: exploring potential mechanisms.bioRxiv [Preprint]. 2024 Oct 27:2024.10.24.619909. doi: 10.1101/2024.10.24.619909. bioRxiv. 2024. Update in: Alzheimers Dement. 2025 Jul;21(7):e70500. doi: 10.1002/alz.70500. PMID: 39484566 Free PMC article. Updated. Preprint.
References
MeSH terms
Substances
Grants and funding
- U01 AG068057/AG/NIA NIH HHS/United States
- U01 AG049505/AG/NIA NIH HHS/United States
- P20 AG068082/AG/NIA NIH HHS/United States
- P30 AG072975/AG/NIA NIH HHS/United States
- R01 AG042188/AG/NIA NIH HHS/United States
- P30 AG066444/AG/NIA NIH HHS/United States
- RC2 HL102419/HL/NHLBI NIH HHS/United States
- N01 HC085080/HL/NHLBI NIH HHS/United States
- R01 AG064877/AG/NIA NIH HHS/United States
- R01 AG018016/AG/NIA NIH HHS/United States
- R01 AG060747/AG/NIA NIH HHS/United States
- P30 AG066507/AG/NIA NIH HHS/United States
- P30 AG072946/AG/NIA NIH HHS/United States
- U01 AG058654/AG/NIA NIH HHS/United States
- U01 HL096812/HL/NHLBI NIH HHS/United States
- U54 AG052427/AG/NIA NIH HHS/United States
- R01 NS017950/NS/NINDS NIH HHS/United States
- P30 AG066518/AG/NIA NIH HHS/United States
- R01 AG019771/AG/NIA NIH HHS/United States
- R01 AG054076/AG/NIA NIH HHS/United States
- R01AG070864/AG/NIA NIH HHS/United States
- R01 AG046949/AG/NIA NIH HHS/United States
- R01 AG042210/AG/NIA NIH HHS/United States
- U54 HG003067/HG/NHGRI NIH HHS/United States
- R01 AG057909/AG/NIA NIH HHS/United States
- U01 AG058922/AG/NIA NIH HHS/United States
- P20 AG068053/AG/NIA NIH HHS/United States
- R01 AG009029/AG/NIA NIH HHS/United States
- AARFD-24-1309441/Alzheimer's Association Research Fellowship to Promote Diversity
- R01 NS069719/NS/NINDS NIH HHS/United States
- R01 AG015928/AG/NIA NIH HHS/United States
- R01 AG044546/AG/NIA NIH HHS/United States
- P01 NS026630/NS/NINDS NIH HHS/United States
- P30 AG010133/AG/NIA NIH HHS/United States
- U24 AG021886/AG/NIA NIH HHS/United States
- R01 AG009956/AG/NIA NIH HHS/United States
- U01 HL080295/HL/NHLBI NIH HHS/United States
- P50 AG016574/AG/NIA NIH HHS/United States
- P30 AG066511/AG/NIA NIH HHS/United States
- U24 AG072122/AG/NIA NIH HHS/United States
- N01 HC085082/HL/NHLBI NIH HHS/United States
- P30 AG066512/AG/NIA NIH HHS/United States
- U01 AG049507/AG/NIA NIH HHS/United States
- U01 HL096917/HL/NHLBI NIH HHS/United States
- U01 AG052411/AG/NIA NIH HHS/United States
- R01 AG019085/AG/NIA NIH HHS/United States
- U01 AG032984/AG/NIA NIH HHS/United States
- U01AG052410/AG/NIA NIH HHS/United States
- P30 AG066515/AG/NIA NIH HHS/United States
- RF1 AG053303/AG/NIA NIH HHS/United States
- U01 HL130114/HL/NHLBI NIH HHS/United States
- P30 AG062421/AG/NIA NIH HHS/United States
- R01 AG048234/AG/NIA NIH HHS/United States
- U01 AG024904/AG/NIA NIH HHS/United States
- N01 HC085083/HL/NHLBI NIH HHS/United States
- N01 HC085086/HL/NHLBI NIH HHS/United States
- U01 AG072579/AG/NIA NIH HHS/United States
- RF1 AG054074/AG/NIA NIH HHS/United States
- R01AG15819/AG/NIA NIH HHS/United States
- P50 NS039764/NS/NINDS NIH HHS/United States
- P30 AG066508/AG/NIA NIH HHS/United States
- P01 AG003991/AG/NIA NIH HHS/United States
- U01 HL096902/HL/NHLBI NIH HHS/United States
- U24 AG056270/AG/NIA NIH HHS/United States
- P30 AG072978/AG/NIA NIH HHS/United States
- P01 AG026276/AG/NIA NIH HHS/United States
- R01 AG017917/AG/NIA NIH HHS/United States
- RF1 AG058501/AG/NIA NIH HHS/United States
- R56 AG051876/AG/NIA NIH HHS/United States
- P30 AG062429/AG/NIA NIH HHS/United States
- P30 AG066519/AG/NIA NIH HHS/United States
- A2018425S/BrightFocus foundation
- R01 AG028786/AG/NIA NIH HHS/United States
- U54 HG003273/HG/NHGRI NIH HHS/United States
- R01 AG049607/AG/NIA NIH HHS/United States
- UF1 AG047133/AG/NIA NIH HHS/United States
- U01 AG057659/AG/NIA NIH HHS/United States
- U24 AG074855/AG/NIA NIH HHS/United States
- R01 AG054047/AG/NIA NIH HHS/United States
- R01 AG061155/AG/NIA NIH HHS/United States
- P50 AG005136/AG/NIA NIH HHS/United States
- P30 AG072973/AG/NIA NIH HHS/United States
- RF1 AG059018/AG/NIA NIH HHS/United States
- R01 AG027161/AG/NIA NIH HHS/United States
- RF1 AG054023/AG/NIA NIH HHS/United States
- R01 HL105756/HL/NHLBI NIH HHS/United States
- P30 AG062422/AG/NIA NIH HHS/United States
- RF1 AG054080/AG/NIA NIH HHS/United States
- U19 AG024904/AG/NIA NIH HHS/United States
- R01 AG079280/AG/NIA NIH HHS/United States
- RF1 AG051504/AG/NIA NIH HHS/United States
- U01 AG062943/AG/NIA NIH HHS/United States
- P30AG066511/AG/NIA NIH HHS/United States
- U01AG066767/AG/NIA NIH HHS/United States
- RF1 AG058066/AG/NIA NIH HHS/United States
- U24 AG041689/AG/NIA NIH HHS/United States
- R01 AG044829/AG/NIA NIH HHS/United States
- P30 AG066462/AG/NIA NIH HHS/United States
- P30 AG010161/AG/NIA NIH HHS/United States
- P30 AG066530/AG/NIA NIH HHS/United States
- R01 AG033193/AG/NIA NIH HHS/United States
- R01 AG032990/AG/NIA NIH HHS/United States
- R37 AG015473/AG/NIA NIH HHS/United States
- N01 HC055222/HL/NHLBI NIH HHS/United States
- U01 AG058589/AG/NIA NIH HHS/United States
- U01 NS041588/NS/NINDS NIH HHS/United States
- R01 NS080820/NS/NINDS NIH HHS/United States
- R01 AG059716/AG/NIA NIH HHS/United States
- R01 AG072547/AG/NIA NIH HHS/United States
- U01 AG049508/AG/NIA NIH HHS/United States
- P50 AG008012/AG/NIA NIH HHS/United States
- N01 HC085079/HL/NHLBI NIH HHS/United States
- U01 HL096814/HL/NHLBI NIH HHS/United States
- P30 AG066509/AG/NIA NIH HHS/United States
- U01AG072579/AG/NIA NIH HHS/United States
- U01 AG052410/AG/NIA NIH HHS/United States
- U01 AG006781/AG/NIA NIH HHS/United States
- R01 AG041797/AG/NIA NIH HHS/United States
- K23 AG030944/AG/NIA NIH HHS/United States
- RC4 AG039085/AG/NIA NIH HHS/United States
- P20 AG068077/AG/NIA NIH HHS/United States
- U01 AG062602/AG/NIA NIH HHS/United States
- R01 NS029993/NS/NINDS NIH HHS/United States
- P30 AG066546/AG/NIA NIH HHS/United States
- R01 AG064614/AG/NIA NIH HHS/United States
- R01 AG021547/AG/NIA NIH HHS/United States
- P30 AG038072/AG/NIA NIH HHS/United States
- R01 AG032289/AG/NIA NIH HHS/United States
- R01 AG048927/AG/NIA NIH HHS/United States
- U54 HG003079/HG/NHGRI NIH HHS/United States
- R01 AG019757/AG/NIA NIH HHS/United States
- U01 AG052409/AG/NIA NIH HHS/United States
- R01 AG033040/AG/NIA NIH HHS/United States
- RF1 AG054052/AG/NIA NIH HHS/United States
- R01 AG022018/AG/NIA NIH HHS/United States
- U01 AG046139/AG/NIA NIH HHS/United States
- P30 AG072977/AG/NIA NIH HHS/United States
- R01 AG020098/AG/NIA NIH HHS/United States
- P30 AG062677/AG/NIA NIH HHS/United States
- R01 HL070825/HL/NHLBI NIH HHS/United States
- Zenith Award
- R01 AG027944/AG/NIA NIH HHS/United States
- P20 AG068024/AG/NIA NIH HHS/United States
- P30 AG072958/AG/NIA NIH HHS/United States
- R01 AG025259/AG/NIA NIH HHS/United States
- RF1 AG057519/AG/NIA NIH HHS/United States
- U01 HL096899/HL/NHLBI NIH HHS/United States
- P30 AG062715/AG/NIA NIH HHS/United States
- RF1 AG015473/AG/NIA NIH HHS/United States
- U01 AG049506/AG/NIA NIH HHS/United States
- P30 AG066506/AG/NIA NIH HHS/United States
- P30 AG072976/AG/NIA NIH HHS/United States
- WT_/Wellcome Trust/United Kingdom
- R01 AG011101/AG/NIA NIH HHS/United States
- P30 AG066468/AG/NIA NIH HHS/United States
- R01 AG021136/AG/NIA NIH HHS/United States
- U01 AG066767/AG/NIA NIH HHS/United States
- P30 AG072931/AG/NIA NIH HHS/United States
- P30 AG072947/AG/NIA NIH HHS/United States
- U01 AG058635/AG/NIA NIH HHS/United States
- P30 AG072972/AG/NIA NIH HHS/United States
- R01 AG023629/AG/NIA NIH HHS/United States
- P30 AG066514/AG/NIA NIH HHS/United States
- U19 AG066567/AG/NIA NIH HHS/United States
- R01AG072547/AG/NIA NIH HHS/United States
- P30 AG072959/AG/NIA NIH HHS/United States
- R01 AG031272/AG/NIA NIH HHS/United States
- R01 AG011380/AG/NIA NIH HHS/United States
- P50 NS071674/NS/NINDS NIH HHS/United States
- N01 HC085081/HL/NHLBI NIH HHS/United States
- R01 AG070864/AG/NIA NIH HHS/United States
- P30 AG072979/AG/NIA NIH HHS/United States
- RF1 AG058267/AG/NIA NIH HHS/United States
- R01 AG015819/AG/NIA NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

