The mammalian protein MTCH1 can function as an insertase
- PMID: 40704594
- PMCID: PMC12401536
- DOI: 10.1242/jcs.263736
The mammalian protein MTCH1 can function as an insertase
Abstract
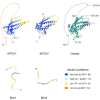
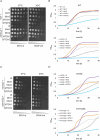
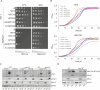
The outer mitochondrial membrane (OMM) hosts a variety of proteins such as import machineries, enzymes, fission and fusion factors, and pore proteins. In Saccharomyces cerevisiae, the MIM complex, consisting of Mim1 and Mim2, mediates the insertion of α-helical proteins into the OMM. Until recently, it was unclear which proteins served this function in higher eukaryotes. Recent studies have identified MTCH2 as the insertase responsible for inserting α-helical proteins into the OMM in mammals. MTCH1 is a paralogue of MTCH2 but its general function and contribution to the biogenesis process are not clear. To better characterize MTCH1, we explored whether MTCH1 or MTCH2 could functionally replace Mim1 and/or Mim2 in yeast. Expression of MTCH1 and MTCH2 in yeast cells lacking Mim1, Mim2 or both revealed that MTCH1, but not MTCH2, could compensate for the growth defects upon deleting the MIM complex. Furthermore, MTCH1 could restore the biogenesis of MIM substrates, translocase of the outer membrane (TOM) complex stability and morphology of mitochondria. These findings indicate that MTCH1, by itself, has insertase activity and is a functional equivalent for the MIM complex, despite the absence of any evolutionary relation between the mammalian and yeast insertases.
Keywords: Insertase; MIM; MTCH1; MTCH2; Mitochondria; Outer membrane.
© 2025. Published by The Company of Biologists.
Conflict of interest statement
Competing interests The authors declare no competing or financial interests.
Figures


References
-
- Becker, T., Wenz, L. S., Krüger, V., Lehmann, W., Müller, J. M., Goroncy, L., Zufall, N., Lithgow, T., Guiard, B., Chacinska, A.et al. (2011). The mitochondrial import protein Mim1 promotes biogenesis of multispanning outer membrane proteins. J. Cell Biol. 194, 387-395. 10.1083/jcb.201102044 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

