Application of Mendelian randomization and bioinformatic analysis to construct a prognostic model for thyroid cancer and perform pan-cancer analysis
- PMID: 40705138
- PMCID: PMC12290152
- DOI: 10.1007/s12672-025-03222-7
Application of Mendelian randomization and bioinformatic analysis to construct a prognostic model for thyroid cancer and perform pan-cancer analysis
Abstract
Objective: This study aimed to identify causal effects and potential molecular mechanisms of genes associated with THCA development.
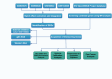
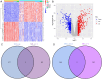
Methods: Bioinformatic analyses were performed to identify differentially expressed genes (DEGs) associated with THCA. Subsequently, Mendelian randomization (MR) analysis was conducted using large-scale eQTL data and THCA GWAS summary statistics to screen for candidate genes. The intersection of DEGs and MR-derived candidate genes was used to determine DEGs with potential causal associations with thyroid carcinogenesis. Functional enrichment analysis, pathway analysis, and immune cell infiltration profiling were performed. External datasets were used for validation. Additionally, prognostic modeling and pan-cancer analyses of the candidate genes were conducted.
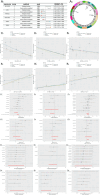
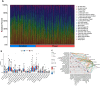
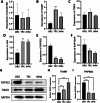
Results: IVW-based MR analysis revealed that elevated expression levels of ALOX15B [OR = 1.647, 95% CI (1.120-2.420), P < 0.05], TIAM1 [OR = 1.270, 95% CI (1.001-1.611), P < 0.05], and TMC6 [OR = 1.250, 95% CI (1.021-1.530), P < 0.05] were associated with an increased risk of THCA. Conversely, elevated expression of JUN [OR = 0.795, 95% CI (0.653-0.967), P < 0.05], PAPSS2 [OR = 0.779, 95% CI (0.608-1.000), P < 0.05], and RAP1GAP [OR = 0.895, 95% CI (0.810-0.989), P < 0.05] was associated with a reduced risk. Gene set enrichment analysis (GSEA) indicated that risk genes were enriched in proliferation- and metastasis-related pathways, such as extracellular matrix (ECM)-receptor interaction and cell adhesion molecules (CAMs). Findings from the training set were further validated experimentally and via external datasets. Additionally, candidate risk genes demonstrated associations with the development and progression of multiple tumor types.
Conclusion: This study identified ALOX15B, TIAM1, and TMC6 as potential risk genes and JUN, PAPSS2, and RAP1GAP as protective genes in THCA. These genes may serve as promising biomarkers and therapeutic targets for THCA, offering novel insights into precision oncology.
Keywords: Bioinformatics; Immune infiltration; Mendelian randomization; Prognostic model; Thyroid cancer.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests.
Figures



References
-
- Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. Cancer J Clin. 2018;68(6):394–424. - PubMed
-
- Dotinga M, Vriens D, van Velden FHP, Stam MK, Heemskerk JWT, Dibbets-Schneider P, Pool M, Rietbergen DDD, de Geus-Oei LF, Kapiteijn E. Reinducing Radioiodine-Sensitivity in Radioiodine-Refractory thyroid Cancer using lenvatinib (RESET): study protocol for a Single-Center, open label phase II trial. Diagnostics (Basel Switzerland) 2022, 12(12). - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
