Identification of key targets and exploration of therapeutic molecular mechanisms of natural compound tangeretin in osteosarcoma
- PMID: 40705154
- PMCID: PMC12290169
- DOI: 10.1007/s12672-025-03221-8
Identification of key targets and exploration of therapeutic molecular mechanisms of natural compound tangeretin in osteosarcoma
Abstract
Introduction: Osteosarcoma (OS) is an invasive and lethal malignancy showing a low 5 year survival rate, underscoring the need for identifying new therapeutic targets and their inhibitors to enhance prevention and treatment strategies.
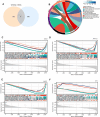
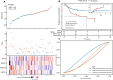
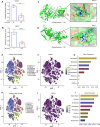
Methods: In this study, in vitro experiments including CCK-8 assay, anchorage-independent growth assays, and plate cloning assays were used to detect the anti-proliferation ability of natural compound tangeretin towards OS cells. An integrated approach was performed including WGCNA and network pharmacology to identify the key genes of tangeretin for the treatment of OS. Multigene diagnostic model, reverse transcription quantitative polymerase chain reaction (RT-qPCR) analysis along with molecular docking analysis were further conducted to validate the reliability of the targets obtained by bioinformatics methods. Single-cell and gene enrichment analyses were chosen to explore the mechanism of tangeretin in OS.
Results: Hub genes identified by the bioinformatics strategy included ABCC1, AKR1C3, BACE1, and CA12. RT-qPCR validation and molecular docking analysis confirmed that ABCC1 and BACE1 were the most likely potential targets. A multigene diagnostic model for OS demonstrated moderate accuracy of the hub genes. Single-cell sequencing results indicated that these two hub targets were closely related to OS and provided more potential mechanisms for targeting OS.
Conclusion: Our research highlights the therapeutic potential of the natural compound tangeretin and its antineoplastic mechanisms in OS. It offers new insights into the molecular mechanisms of tangeretin, paving the way for the development of effective OS treatments.
Keywords: Natural compound; Network pharmacology; Osteosarcoma; Tangeretin; Therapeutic targets; WGCNA.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Not applicable. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures





Similar articles
-
Understanding mechanisms of Polygonatum sibiricum-derived exosome-like nanoparticles against breast cancer through an integrated metabolomics and network pharmacology analysis.Front Chem. 2025 Jun 6;13:1559758. doi: 10.3389/fchem.2025.1559758. eCollection 2025. Front Chem. 2025. PMID: 40547857 Free PMC article.
-
Integrated bioinformatics and network pharmacology to identify and validate macrophage polarization related hub genes in the treatment of osteoarthritis with Astragalus membranaceus.J Orthop Surg Res. 2025 May 30;20(1):543. doi: 10.1186/s13018-025-05799-9. J Orthop Surg Res. 2025. PMID: 40442788 Free PMC article.
-
Elucidating the Mechanism of Xiaoqinglong Decoction in Chronic Urticaria Treatment: An Integrated Approach of Network Pharmacology, Bioinformatics Analysis, Molecular Docking, and Molecular Dynamics Simulations.Curr Comput Aided Drug Des. 2025 Jul 16. doi: 10.2174/0115734099391401250701045509. Online ahead of print. Curr Comput Aided Drug Des. 2025. PMID: 40676786
-
Signs and symptoms to determine if a patient presenting in primary care or hospital outpatient settings has COVID-19.Cochrane Database Syst Rev. 2022 May 20;5(5):CD013665. doi: 10.1002/14651858.CD013665.pub3. Cochrane Database Syst Rev. 2022. PMID: 35593186 Free PMC article.
-
Management of urinary stones by experts in stone disease (ESD 2025).Arch Ital Urol Androl. 2025 Jun 30;97(2):14085. doi: 10.4081/aiua.2025.14085. Epub 2025 Jun 30. Arch Ital Urol Androl. 2025. PMID: 40583613 Review.
References
-
- Sung H, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–49. - PubMed
-
- Bernthal NM, et al. Long-term results (>25 years) of a randomized, prospective clinical trial evaluating chemotherapy in patients with high-grade, operable osteosarcoma. Cancer. 2012;118(23):5888–93. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
