Cell type-specific purifying selection of synonymous mitochondrial DNA variation
- PMID: 40705423
- PMCID: PMC12318227
- DOI: 10.1073/pnas.2505704122
Cell type-specific purifying selection of synonymous mitochondrial DNA variation
Abstract
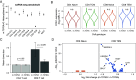
While somatic variants are well-characterized drivers of tumor evolution, their influence on cellular fitness in nonmalignant contexts remains understudied. We identified a mosaic synonymous variant (m.7076A > G) in the mitochondrial DNA (mtDNA)-encoded cytochrome c-oxidase subunit 1 (MT-CO1, p.Gly391=), present at homoplasmy in 47% of immune cells from a healthy donor. Single-cell multiomics revealed strong, lineage-specific selection against the m.7076G allele in CD8+ effector memory T cells, but not other T cell subsets, mirroring patterns of purifying selection of pathogenic mtDNA alleles. The limited anticodon diversity of mitochondrial tRNAs forces m.7076G translation to rely on wobble pairing, unlike the Watson-Crick-Franklin pairing used for m.7076A. Mitochondrial ribosome profiling confirmed stalled translation of the m.7076G allele. Functional analyses demonstrated that the elevated translational and metabolic demands of short-lived effector T cells (SLECs) amplify dependence on MT-CO1, driving this selective pressure. These findings suggest that synonymous variants can alter codon syntax, impacting mitochondrial physiology in a cell type-specific manner.
Keywords: immunology; mitochondria; selection; single-cell.
Conflict of interest statement
Competing interests statement:A.T.S. is a founder of Immunai, Cartography Biosciences, Santa Ana Bio, and Arpelos Biosciences, and receives research funding from Allogene Therapeutics and Merck Research Laboratories. C.A.L. is a consultant for Cartography Biosciences. Stanford University has filed a provisional patent based on this work where C.A.L. and A.T.S. are named inventors. The Broad Institute has filed for patents relating to the use of mtDNA sequencing technologies described in this paper where C.A.L. and L.S.L, are named inventors (US patent applications 17/251,451 and 17/928,696).
Figures




References
-
- Stewart J. B., Chinnery P. F., The dynamics of mitochondrial DNA heteroplasmy: Implications for human health and disease. Nat. Rev. Genet. 16, 530–542 (2015). - PubMed
-
- Yazar S., et al. , Single-cell eQTL mapping identifies cell type–specific genetic control of autoimmune disease. Science 376, eabf3041 (2022). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

