Lyophilised Reverse Transcriptase and Polymerase for Point-of-Care Diagnostics of SARS-CoV-2
- PMID: 40705463
- PMCID: PMC12288692
- DOI: 10.1111/1751-7915.70200
Lyophilised Reverse Transcriptase and Polymerase for Point-of-Care Diagnostics of SARS-CoV-2
Abstract
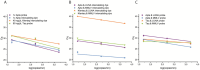
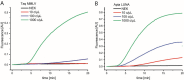
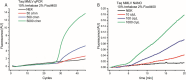
Early diagnosis of pathogens is key to reducing their spreading and to preventing severe health risks. The COVID-19 pandemic showed us the need for rapid point-of-care tests. Here, we describe the preparation of an amplification master mix for point-of-care diagnostics. Therefore, two off-patent amplification enzymes were designed, expressed and purified. The preparation of the key components enables independence from delivery issues, manufacturer portfolio and product information's. For long-term storage and cold-free transport, our fabricated amplification mix was lyophilised. Finally, we applied our lyophilised master mix on an integrated point-of-care diagnostic system and could detect 10 copies/μL COVID-19 RNA. The combination of stable, cold-free reagents with the mobile and low-cost device will allow molecular diagnostics of pathogens in a field or home setting.
Keywords: RT‐qPCR; SARS‐CoV‐2; lyophilisation; point‐of‐care; pulse controlled amplification; recombinant enzymes.
© 2025 The Author(s). Microbial Biotechnology published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

References
-
- Barnes, W. M. 1992. “The Fidelity of Taq Polymerase Catalyzing PCR Is Improved by an N‐Terminal Deletion.” Gene 112: 29–35. - PubMed
-
- Chen, Y. , Xu W., and Sun Q.. 2009. “A Novel and Simple Method for High‐Level Production of Reverse Transcriptase From Moloney Murine Leukemia Virus (MMLV‐RT) in <styled-content style="fixed-case"> Escherichia coli </styled-content> .” Biotechnology Letters 31: 1051–1057. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

