A predictive endocrine resistance index accurately stratifies luminal breast cancer treatment responders and nonresponders
- PMID: 40705465
- PMCID: PMC12483570
- DOI: 10.1172/JCI177813
A predictive endocrine resistance index accurately stratifies luminal breast cancer treatment responders and nonresponders
Abstract
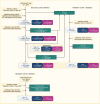
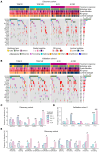
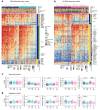
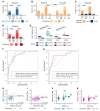
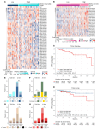
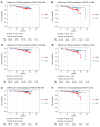
BACKGROUNDEndocrine therapy (ET) with tamoxifen (TAM) or aromatase inhibitors (AI) is highly effective against hormone receptor-positive (HR-positive) early breast cancer (BC), but resistance remains a major challenge. The primary objectives of our study were to understand the underlying mechanisms of primary resistance and to identify potential biomarkers.METHODSWe selected more than 800 patients in 3 subcohorts (Discovery, n = 364, matched pairs; Validation 1, n = 270, Validation 2, n = 176) of the West German Study Group (WSG) ADAPT trial who underwent short-term preoperative TAM or AI treatment. Treatment response was assessed by immunohistochemical labeling of proliferating cells with Ki67 before and after ET. We performed comprehensive molecular profiling, including targeted next-generation sequencing (NGS) and DNA methylation analysis using EPIC arrays, on posttreatment tumor samples.RESULTSTP53 mutations were strongly associated with primary resistance to both TAM and AI. We identified distinct DNA methylation patterns in resistant tumors, suggesting alterations in key signaling pathways and tumor microenvironment composition. Based on these findings and patient age, we developed the Predictive Endocrine ResistanCe Index (PERCI). PERCI accurately stratified responders and nonresponders in both treatment groups in all 3 subcohorts and predicted progression-free survival in an external validation cohort and in the combined subcohorts.CONCLUSIONOur results highlight the potential of PERCI to guide personalized endocrine therapy and improve patient outcomes.TRIAL REGISTRATIONWSG-ADAPT, ClinicalTrials.gov NCT01779206, retrospectively registered 01-25-2013.FUNDINGGerman Cancer Aid (Grant Number 70112954), German Federal Ministry of Education and Research (Grant Number 01ZZ1804C, DIFUTURE).
Keywords: Bioinformatics; Breast cancer; Clinical Research; Clinical trials; Epigenetics; Oncology.
Conflict of interest statement
Figures


References
Publication types
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

