Protocol for the establishment and characterization of South African patient-derived intestinal organoids
- PMID: 40705597
- PMCID: PMC12305567
- DOI: 10.1016/j.xpro.2025.103970
Protocol for the establishment and characterization of South African patient-derived intestinal organoids
Abstract
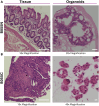
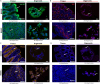
Patient-derived organoids are valuable for modeling disease pathogenesis, therapeutic screening, and advancing personalized medicine. Here, we present a protocol for generating and characterizing intestinal organoids from South African patients, using both healthy and cancerous tissues. We describe steps for generating organoid cultures, quantitative reverse-transcription polymerase chain reaction (RT-qPCR), immunofluorescence microscopy, and histological staining. Notably, we provide a detailed method for cholesterol profiling in organoids and tissues, offering a powerful tool for researchers exploring lipid biology in organoid systems.
Keywords: Cancer; Molecular Biology; Organoids.
Copyright © 2025 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures














References
-
- Sato T., Stange D.E., Ferrante M., Vries R.G.J., van Es J.H., van den Brink S., van Houdt W.J., Pronk A., van Gorp J., Siersema P.D., Clevers H. Long-term Expansion of Epithelial Organoids From Human Colon, Adenoma, Adenocarcinoma, and Barrett's Epithelium. Gastroenterology. 2011;141:1762–1772. doi: 10.1053/j.gastro.2011.07.050. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

