Untargeted Diversity-Oriented Synthesis for the Discovery of New Antitumor Agents: An Integrated Approach of Inverse Virtual Screening, Bioinformatics, and Omics for Target Deconvolution
- PMID: 40705771
- PMCID: PMC12362587
- DOI: 10.1021/acs.jmedchem.5c01344
Untargeted Diversity-Oriented Synthesis for the Discovery of New Antitumor Agents: An Integrated Approach of Inverse Virtual Screening, Bioinformatics, and Omics for Target Deconvolution
Abstract
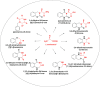
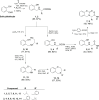
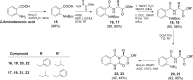
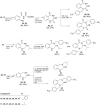
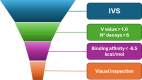
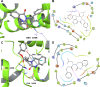
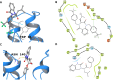
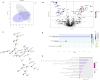
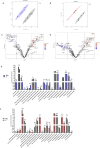
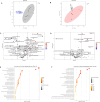
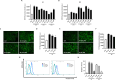
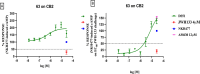
Molecular diversity is one of the most pursued objectives in drug discovery, and diversity-oriented synthesis (DOS) perfectly responds to the achievement of this goal. In this paper, we describe a DOS approach applied to the antitumor field with the aim of identifying new anticancer structures and their associated targets. To accomplish this ambitious project, after an initial stage of phenotypic evaluation, we set up an integrated platform of inverse virtual screening (IVS), bioinformatics, and omics to predict the biological targets of the most promising compounds 31 and 63. Several proteins emerged from this study, and the most interesting ones were assessed by biophysical and in cellulo experiments, leading to the validation of six targets involved in calcium regulation, endoplasmic reticulum stress, and apoptosis. This work allowed us to identify two hit compounds with an interesting antitumor mechanism, but principally, to validate our platform as a fruitful tool for untargeted DOS campaigns.
Figures



References
-
- Sinha S., Vohora D.. Drug Discovery and Development. In Pharmaceutical Medicine and Translational Clinical Research. 2018:19–32. doi: 10.1016/B978-0-12-802103-3.00002-X. - DOI
-
- Lood C. S., Koskinen A. M. P.. Harmicine, a Tetracyclic Tetrahydro-β-Carboline: From the First Synthetic Precedent to Isolation from Natural Sources to Target-Oriented Synthesis (Review)*. Chem. Heterocycl. Compd. 2015;50(10):1367–1387. doi: 10.1007/s10593-014-1602-4. - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

