Catalytic Mechanism of the Bacterial Non-Heme Fe(II) and 2-Oxoglutarate Dependent Enzyme AlkB with Single-Stranded DNA Containing Complex Guanine Adducts
- PMID: 40706026
- PMCID: PMC12380083
- DOI: 10.1021/acs.inorgchem.5c02176
Catalytic Mechanism of the Bacterial Non-Heme Fe(II) and 2-Oxoglutarate Dependent Enzyme AlkB with Single-Stranded DNA Containing Complex Guanine Adducts
Abstract
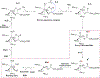
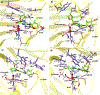
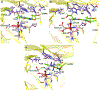
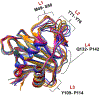
The bacterial nonheme Fe(II)/2-oxoglutarate (2OG)-dependent enzyme AlkB repairs alkylation damages in single-stranded DNA (ss-DNA) nucleotide bases. This study examines for the first time the reaction mechanism of the AlkB-catalyzed repair of alkylated and exocyclic guanine adducts (GAs) in single-stranded DNA induced by everyday chemical exposures associated with cancers and other genetic disorders. The studied substrates include N2-furfurylguanine (FF-dG), N2-tetrahydrofuran-2-yl-methylguanine (HF-dG), 3-(2'-deoxy-β-D-erythro-pentofuranosyl)-5,6,7,8-tetrahydro-6-hydroxypyrimido[1,2-α]purin-10(3H)-one (α-OH-PdG), 3-(2'-deoxy-β-D-erythro-pentofuranosyl)-5,6,7,8-tetrahydro-8-hydroxypyrimido[1,2-α]purin-10(3H)-one (γ-OH-PdG), and 3-(2'-deoxy-β-D-erythro-pentofuranosyl) pyrimido[1,2-α]purin-10(3H)-one (M1dG). Using molecular dynamics-based combined quantum mechanics/molecular mechanics (QM/MM) and QM calculations, we provide unique mechanistic insights into AlkB's catalytic reaction pathways with ss-DNA containing complex alkylated/exocyclic GAs in strong correlation to experimental studies. While HF-dG, FF-dG, α-OH-PdG, and γ-OH-PdG are repaired through C-H hydroxylation, M1dG follows epoxidation. The study elucidated that the repair mechanism favors the open tautomer of γ-OH-PdG and the closed tautomer of α-OH-PdG, respectively, in agreement with experimental studies, due to the preferable SCS interactions and the catalytic domain's loop L1 and L4 dynamics. Our study also elucidated that the posthydroxylation/postepoxidation steps proceed in water rather than the enzyme. The results reveal the unique catalytic mechanism of AlkB with ss-DNA containing complex GAs, which can be used in drug design and metalloenzyme redesign.
Figures






 .
.




References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

