HLA-E[pHLA-G] complex-specific monoclonal antibody enhancing NK activity in multiple myeloma
- PMID: 40706037
- PMCID: PMC12596974
- DOI: 10.1182/bloodadvances.2025016276
HLA-E[pHLA-G] complex-specific monoclonal antibody enhancing NK activity in multiple myeloma
Abstract
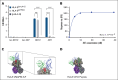
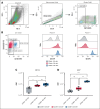
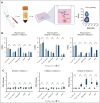
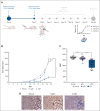
HLA-E presenting the HLA-G leader peptide VMAPRTLFL (HLA-E[pHLA-G]) on tumor cells plays a crucial role in suppressing natural killer (NK) and cytotoxic CD8+ T cells through NKG2A interaction. While blocking HLA-E:NKG2A is a promising immune checkpoint (IC) approach in cancer therapy, toxicity remains a major clinical concern. We developed a novel IC inhibitor that selectively prevents HLA-E:NKG2A interaction, a monoclonal antibody that selectively targets the HLA-E[pHLA-G] complex, distinguishing cancerous from noncancerous cells. In clinical bone marrow samples from patients with multiple myeloma (MM), 4D7 specifically recognized tumor-associated HLA-E-peptide complexes. Using NK cells from healthy donors, 4D7 effectively blocked the HLA-E:NKG2A interaction, and enhanced NKG2A-positive NK cell activity in autologous MM cell cocultures. Importantly, 4D7 did not inhibit NKG2C-positive NK cells, preserving their activity, even though NKG2C also interacts with HLA-E. In MM-bearing mice treated with human NK cells, 4D7 significantly reduced tumor growth. This targeted approach activates NK cells only against tumor cells presenting HLA-E-peptide complexes, potentially minimizing toxicity compared with current NKG2A inhibitors. The development of 4D7 highlights a promising advancement in immunotherapy for hematologic malignancies, offering improved outcomes for patients with MM, and a foundation for broader application across cancer types.
© 2025 American Society of Hematology. Published by Elsevier Inc. Licensed under Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International (CC BY-NC-ND 4.0), permitting only noncommercial, nonderivative use with attribution. All other rights reserved.
Conflict of interest statement
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Figures



References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

