Ether lipids and sphingolipids drive sex-specific human aging dynamics
- PMID: 40706291
- PMCID: PMC12309944
- DOI: 10.1016/j.redox.2025.103779
Ether lipids and sphingolipids drive sex-specific human aging dynamics
Abstract
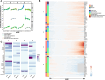
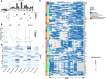
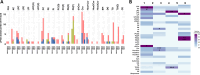
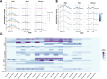
Aging is a dynamic process characterized by complex molecular changes, including shifts in lipid metabolism. To systematically define lipidome dynamics with age and identify sex-specific lipidomic signatures, we performed targeted lipidomic profiling of plasma samples from 1030 adults aged 50-98 years, analyzing 543 lipid species across all lipid classes using high-throughput mass spectrometry and assessing the circulating fatty acid composition by gas chromatography. Our results reveal age-related lipidomic shifts, with ceramides and ether-linked phospholipids most affected. We identified three aging crests (55-60, 65-70, 75-80 years), with the 65-70 years crest dominant in men and the 75-80 years crest in women. Lipid enrichment analyses highlight acylcarnitines, sphingolipids and ether-linked phospholipids as key contributors, with functional indices indicating compositional shifts in lipid species. These findings suggest an impairment of lipid functional categories, including loss of dynamic properties, alterations in bioenergetics, antioxidant defense, cellular identity, and signaling platforms. This study underscores the non-linear nature of lipid metabolism in aging and provides a foundation for identifying biomarkers and interventions to promote healthy aging.
Keywords: Aging crests; Aging dynamics; Ether-linked phospholipids; Lipid metabolism; Metabolic adaptation; Sex-specific lipidomics; Sphingolipids.
Copyright © 2025 The Authors. Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of competing interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures




Similar articles
-
Lipidomic Profiling of Red Blood Cells in the Mitochondrial Fatty Acid β-oxidation Disorder MCADD Reveals Phospholipid and Sphingolipid Dysregulation.J Proteome Res. 2025 Jul 24. doi: 10.1021/acs.jproteome.5c00308. Online ahead of print. J Proteome Res. 2025. PMID: 40704861
-
Non-Targeted Lipidomics Analysis of Characteristic Milk Using High-Resolution Mass Spectrometry (UHPLC-HRMS).Foods. 2025 Jun 12;14(12):2068. doi: 10.3390/foods14122068. Foods. 2025. PMID: 40565677 Free PMC article.
-
Metabolic lipid alterations in subclinical ketotic dairy cows: A multisample lipidomic approach.J Dairy Sci. 2025 Aug;108(8):8887-8903. doi: 10.3168/jds.2025-26442. Epub 2025 Jul 16. J Dairy Sci. 2025. PMID: 40675483
-
Lipidomic Expression Analysis in Carotid Atherosclerotic Disease: A Systematic Review.Ann Vasc Surg. 2025 Apr;113:83-94. doi: 10.1016/j.avsg.2024.12.063. Epub 2025 Jan 22. Ann Vasc Surg. 2025. PMID: 39855383
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2021 Apr 19;4(4):CD011535. doi: 10.1002/14651858.CD011535.pub4. Cochrane Database Syst Rev. 2021. Update in: Cochrane Database Syst Rev. 2022 May 23;5:CD011535. doi: 10.1002/14651858.CD011535.pub5. PMID: 33871055 Free PMC article. Updated.
References
-
- Sol J., Ortega-Bravo M., Portero-Otín M., Piñol-Ripoll G., Ribas-Ripoll V., Artigues-Barberà E., Butí M., Pamplona R., Jové M. Human lifespan and sex-specific patterns of resilience to disease: a retrospective population-wide cohort study. BMC Med. 2024;22 doi: 10.1186/S12916-023-03206-W. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

