Rapid, sensitive, and species-independent detection of Crimean Congo hemorrhagic fever virus nucleoprotein and GP38 antibodies
- PMID: 40706445
- PMCID: PMC12305605
- DOI: 10.1016/j.ebiom.2025.105857
Rapid, sensitive, and species-independent detection of Crimean Congo hemorrhagic fever virus nucleoprotein and GP38 antibodies
Abstract
Background: Crimean-Congo hemorrhagic fever virus (CCHFV), a zoonotic agent in the Nairoviridae family (genus Orthonairovirus), is a high-priority pathogen. CCHFV infection causes Crimean-Congo hemorrhagic fever (CCHF), a human disease with case fatality rates of up to 40%. Serological surveillance of CCHFV in animals and humans is crucial for ecological studies and public health.
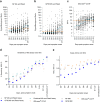
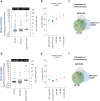
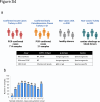
Methods: We developed CCHFV mix-and-read assays utilizing split-NanoLuc technology (NanoBiT) to detect anti-CCHFV antibodies against the nucleoprotein (NP) stalk region and the GP38 glycoprotein. These species- and isotype-agnostic assays provide results in ∼30 min. Using serum samples from RT-PCR-confirmed CCHF cases collected during and after hospitalization, we investigated anti-NP and anti-GP38 antibody development. The performance of the mix-and-read assays was compared to the NP-based IDScreen® CCHF commercial assay using human sera, and cross-reactivity potential was evaluated using a diverse panel of anti-orthonairovirus antisera raised in mice.
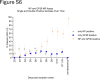
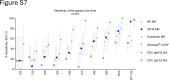
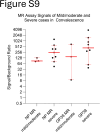
Findings: In human convalescent cases (n = 21), mix-and-read assay concordance between anti-GP38 and anti-NP antibody detection was 100%. Both mix-and-read assays and IDScreen® CCHF demonstrated identical sensitivity of 95.2% in convalescent patients. The specificity of the NP assay was 98.9%, and that of GP38 was 99.7%, both comparable to IDScreen® CCHF (specificity: 99.7%). Cross-reactivity against CCHF NP and GP38, regardless of assay type, was primarily observed in antisera raised against other orthonairoviruses within the Nairobi sheep disease genogroup.
Interpretation: The simplicity and robust performance of the CCHFV mix-and-read assays make them ideal tools for supporting serological surveillance in humans and animals. Furthermore, the inclusion of the GP38 antigen alongside NP enhances the precise identification of retrospective CCHF cases, further strengthening broad surveillance efforts.
Funding: CDC Emerging Infectious Disease Research Core Funds, funding for reagent, CDC personal, travel. Defence Threat Reduction Agency (HDTRA12210007): E.K. salary. Oak Ridge Institute for Science and Education (ORISE): E.K. salary and travel. National Institute of Allergy and Infectious Diseases (1R01AI180125-01A1): sample acquisition. Funding sources did not have a role in the writing or decision to submit the publication.
Keywords: Animal surveillance; Antibody detection; Antibody kinetics; Biosensor; CCHF; Crimean-Congo hemorrhagic fever; Cross-reactivity; Diagnostic techniques; Disease severity; ELISA; GP38; Nucleoprotein; Orthonairovirus; Serological diagnosis; Serosurveillance; Tick-borne diseases.
Published by Elsevier B.V.
Conflict of interest statement
Declaration of interests E.K. was supported by the Oak Ridge Institute for Science and Education (ORISE) fellowship. M.M.S., E.B., and S.D.P. were supported by the National Institute of Allergy and Infectious Diseases (NIAID) grant 1R01AI180125-01A1. E.B. and S.D.P. also received funding from NIAID (1R01AI151006) and the Defence Threat Reduction Agency (HDTRA12210007). The funding sources had no role in the design, writing, or decision to submit this manuscript.
Figures


















References
-
- Dvm P.F., Schnepf G., Gonzalez-Martin F., Bi Z. In: Crimean-Congo hemorrhagic fever. Ergonul O., Whitehouse C.A., editors. 2007. International surveillance and control of Crimean-Congo hemorrhagic fever outbreaks; pp. 295–303. Dordrecht: Springer Netherlands.
-
- Estrada-Peña A., Jameson L., Medlock J., Vatansever Z., Tishkova F. Unraveling the ecological complexities of tick-associated Crimean-Congo hemorrhagic fever virus transmission: a gap analysis for the western Palearctic. Vector Borne Zoonotic Dis. 2012;12(9):743–752. - PubMed
-
- Bente D.A., Forrester N.L., Watts D.M., McAuley A.J., Whitehouse C.A., Bray M. Crimean-Congo hemorrhagic fever: history, epidemiology, pathogenesis, clinical syndrome and genetic diversity. Antiviral Res. 2013;100(1):159–189. - PubMed
-
- W H O . Crimean-Congo Haemorrhagic Fever; 2022. Crimean-Congo haemorrhagic fever.https://www.who.int/news-room/fact-sheets/detail/crimean-congo-haemorrha... [cited 2023 Dec 5]. Available from:
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

