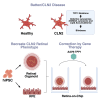
Recreating pathophysiology of CLN2 disease and demonstrating reversion by TPP1 gene therapy in hiPSC-derived retinal organoids and retina-on-chip
- PMID: 40706588
- PMCID: PMC12432371
- DOI: 10.1016/j.xcrm.2025.102244
Recreating pathophysiology of CLN2 disease and demonstrating reversion by TPP1 gene therapy in hiPSC-derived retinal organoids and retina-on-chip
Abstract
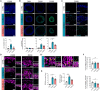
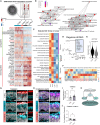
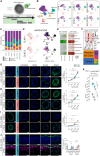
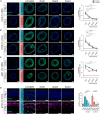
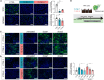
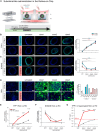
Mutations in the tripeptidyl peptidase 1 (TPP1) gene lead to neuronal ceroid lipofuscinosis type 2 (CLN2), characterized by lysosomal accumulation of lipofuscins predominantly in the brain and retina. The ocular phenotype is characterized by outer retinal degeneration that leads to vision loss. Leveraging human induced pluripotent stem cell (hiPSC)-derived retinal organoids (ROs), retinal pigmented epithelial cells, and the retina-on-chip system, we establish an in vitro CLN2 model that recreates the principal histological hallmarks, namely the accumulation of subunit C of mitochondrial ATP synthase (SCMAS) and lipids mainly in the outer retina. Furthermore, single-cell RNA sequencing reveals a dysregulation of translational and mitochondrial function in CLN2 cones. Finally, adeno-associated virus (AAV)-mediated TPP1 gene therapy can restore TPP1 expression and decrease and even prevent SCMAS accumulations. Our study uses an innovative human-relevant microphysiological retinal disease models, uncovers previously uncharacterized mechanisms of CLN2 pathophysiology, and demonstrates the potential of AAV9.hCLN2 gene therapy for CLN2 disease, potentially treating patient blindness.
Keywords: CLN2; SCMAS; TPP1; gene therapy; neuronal ceroid lipofuscinosis type 2; retina-on-chip; retinal organoids; subunit C of mitochondrial ATP synthase; tripeptidyl peptidase 1.
Copyright © 2025 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests K.H.K., C.B.T., M.H., R.M.R., T.C., and N.B. are current or previous employees of REGENXBIO, Inc. K.H.K. and N.B. are inventors on patent applications related to AAV9.hCLN2 vector delivery methods. N.B. is an inventor on patent applications related to AAV formulations. K.A., S.L., and P. Loskill hold a patent on the RoC technology.
Figures

References
-
- Palmer D.N., Martinus R.D., Cooper S.M., Midwinter G.G., Reid J.C., Jolly R.D. Ovine ceroid lipofuscinosis. The major lipopigment protein and the lipid-binding subunit of mitochondrial ATP synthase have the same NH2-terminal sequence. J. Biol. Chem. 1989;264:5736–5740. - PubMed
-
- Palmer D.N., Fearnley I.M., Medd S.M., Walker J.E., Martinus R.D., Bayliss S.L., Hall N.A., Lake B.D., Wolfe L.S., Jolly R.D. Lysosomal storage of the DCCD reactive proteolipid subunit of mitochondrial ATP synthase in human and ovine ceroid lipofuscinoses. Adv. Exp. Med. Biol. 1989;266:211–223. doi: 10.1007/978-1-4899-5339-1_15. - DOI - PubMed
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical

