IgA2+ B cells and IgA2 anti-dsDNA antibodies are selectively targeted by belimumab after rituximab therapy in systemic lupus erythematosus
- PMID: 40706590
- PMCID: PMC12432362
- DOI: 10.1016/j.xcrm.2025.102247
IgA2+ B cells and IgA2 anti-dsDNA antibodies are selectively targeted by belimumab after rituximab therapy in systemic lupus erythematosus
Abstract
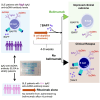
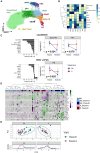
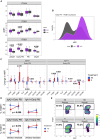
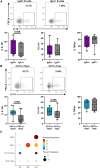
No theragnostic biomarkers exist for systemic lupus erythematosus (SLE) to enable a precision medicine approach. Baseline serum IgA2 anti-double-stranded DNA (dsDNA) antibody levels are associated with response to combination belimumab after rituximab therapy in SLE (BEAT-lupus trial, ISRCTN 47873003). Analysis of the CALIBRATE trial (NCT02260934) confirms that baseline IgA2 anti-dsDNA antibody levels are specifically associated with response to belimumab after rituximab (odds ratio [OR] = 16.9, confidence interval [CI]: 2.8-101, compared to rituximab alone-CALIBRATE and BEAT-lupus combined data). IgA2 anti-dsDNA antibody levels decrease alongside IgA2 expression in plasmablasts only after this combination treatment. Increased serum B cell-activating factor (BAFF) levels are associated with rising IgA2 anti-dsDNA antibody levels after rituximab. IgA2 plasmablasts have increased BAFF receptor and interleukin (IL)-10 expression compared to IgA1 plasmablasts and have a distinct integrin profile implicating a gut mucosal origin. These findings validate IgA2 anti-dsDNA antibodies as a theragnostic biomarker of response and provide mechanistic insight into the selective targeting of IgA2+ B cells by combination belimumab after rituximab in SLE.
Keywords: BAFF; CD11c(+)Tbet(+) age-related B cells; IgA2 anti-dsDNA antibodies; SLE; belimumab; rituximab; theragnostic biomarker.
Copyright © 2025 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests M.R.E. has received grant/research support from GSK. M.R.E. and J.A.J. have received consultancy fees for attending GSK advisory boards. M.R.A.S. and M.R.E. are named on patent application no. 18/864,514 (IgA2 anti-dsDNA antibodies as a biomarker in SLE, the patent is to University College London).
Figures



References
-
- Merrill J.T., Neuwelt C.M., Wallace D.J., Shanahan J.C., Latinis K.M., Oates J.C., Utset T.O., Gordon C., Isenberg D.A., Hsieh H.J., et al. Efficacy and safety of rituximab in moderately-to-severely active systemic lupus erythematosus: the randomized, double-blind, phase II/III systemic lupus erythematosus evaluation of rituximab trial. Arthritis Rheum. 2010;62:222–233. - PMC - PubMed
-
- Rovin B.H., Furie R., Latinis K., Looney R.J., Fervenza F.C., Sanchez-Guerrero J., Maciuca R., Zhang D., Garg J.P., Brunetta P., et al. Efficacy and safety of rituximab in patients with active proliferative lupus nephritis: the Lupus Nephritis Assessment with Rituximab study. Arthritis Rheum. 2012;64:1215–1226. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

