Restriction of Ku translocation protects telomere ends
- PMID: 40707444
- PMCID: PMC12289973
- DOI: 10.1038/s41467-025-61864-1
Restriction of Ku translocation protects telomere ends
Abstract
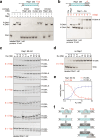
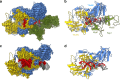
Safeguarding chromosome ends against fusions via nonhomologous end joining (NHEJ) is essential for genome integrity. Paradoxically, the conserved NHEJ core factor Ku binds telomere ends. How it is prevented from promoting NHEJ remains unclear, as does the mechanism that allows Ku to coexist with telomere-protective DNA binding proteins, Rap1 in Saccharomyces cerevisiae. Here, we find that Rap1 directly inhibits Ku's NHEJ function at telomeres. A single Rap1 molecule near a double-stand break suppresses NHEJ without displacing Ku in cells. Furthermore, Rap1 and Ku form a complex on short DNA duplexes in vitro. Cryo-EM shows Rap1 blocks Ku's inward translocation on DNA - an essential step for NHEJ at DSBs. Nanopore sequencing of telomere fusions confirms this mechanism protects native telomere ends. These findings uncover a telomere protection mechanism where Rap1 restricts Ku's inward translocation. This switches Ku from a repair-promoting to a protective role preventing NHEJ at telomeres.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures



References
-
- de Lange, T. Shelterin-mediated telomere protection. Annu. Rev. Genet.52, 223–247 (2018). - PubMed
-
- Chan, S. W.-L. & Blackburn, E. H. Telomerase and ATM/Tel1p protect telomeres from nonhomologous end joining. Mol. Cell11, 1379–1387 (2003). - PubMed
-
- Stuart, A. & De Lange, T. Replicative senescence is ATM driven, reversible, and accelerated by hyperactivation of ATM at normoxia. Preprint at 10.1101/2024.06.24.600514 (2024).
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

