A draft UAE-based Arab pangenome reference
- PMID: 40707445
- PMCID: PMC12290100
- DOI: 10.1038/s41467-025-61645-w
A draft UAE-based Arab pangenome reference
Abstract
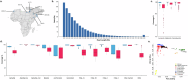
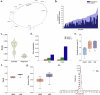
Pangenomes provide a robust and comprehensive portrayal of genetic diversity in humans, but Arab populations remain underrepresented. We present a preliminary UAE-based Arab Pangenome Reference (UPR) utilizing 53 individuals of diverse Arab ethnicities residing in the United Arab Emirates. We assembled nuclear and mitochondrial pangenomes using 35.27X high-fidelity long reads, 54.22X ultralong reads and 65.46X Hi-C reads. This approach yielded contiguous haplotype-phased de novo assemblies of exceptional quality, with an average N50 of 124.28 Mb. We discovered 111.96 million base pairs of previously uncharacterized euchromatic sequences absent from existing human pangenomes, the T2T-CHM13 and GRCh38 reference human genomes, and other public datasets. Moreover, we identified 8.94 million population-specific small variants and 235,195 structural variants within the Arab pangenome, not present in linear and pangenome references and public datasets. We detected 883 gene duplications, including the TATA-binding protein gene TAF11L5, which was uniquely duplicated across all Arab populations and that included 15.06% of genes associated with recessive diseases. By exploring the mitochondrial pangenome, we identified 1,436 bp of previously unreported sequences. Our study provides a valuable resource for future genetic research and genomic medicine initiatives in Arab population and other population with similar genetic backgrounds.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures



References
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

