The A-C linker controls centriole structural integrity and duplication
- PMID: 40707486
- PMCID: PMC12290023
- DOI: 10.1038/s41467-025-62154-6
The A-C linker controls centriole structural integrity and duplication
Abstract
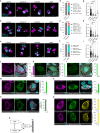
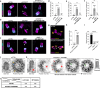
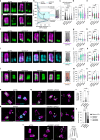
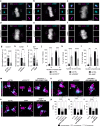
Centrioles are evolutionarily conserved barrel-shaped organelles playing crucial roles in cell division and ciliogenesis. These functions are underpinned by specific structural sub-elements whose functions have been under investigation since many years. The A-C linker structure, connecting adjacent microtubule triplets in the proximal region, has remained unexplored due to its unknown composition. Here, using ultrastructure expansion microscopy, we characterized two recently identified A-C linker proteins, CCDC77 and WDR67, and discovered MIIP as an additional A-C linker protein. Our findings reveal that these proteins localize between microtubule triplets at the A-C linker, forming a complex. Depletion of A-C linker components disrupt microtubule triplet cohesion, leading to breakage at the proximal end. Co-removal of the A-C linker and the inner scaffold demonstrates their joint role in maintaining centriole architecture. Moreover, we uncover an unexpected function of the A-C linker in centriole duplication through torus regulation, underscoring the interplay between these protein modules.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures



References
-
- Nigg, E. A. & Raff, J. W. Centrioles, centrosomes, and cilia in health and disease. Cell139, 663–678 (2009). - PubMed
-
- Doxsey, S. Re-evaluating centrosome function. Nat. Rev. Mol. Cell Biol.2, 688–698 (2001). - PubMed
-
- Bettencourt-Dias, M. & Glover, D. M. Centrosome biogenesis and function: centrosomics brings new understanding. Nat. Rev. Mol. Cell Biol.8, 451–463 (2007). - PubMed
-
- Gönczy, P. Centrosomes and cancer: revisiting a long-standing relationship. 10.1038/nrc3995 (2015). - PubMed
-
- Bornens, M. Centrosome Cycle. Life Sci.521, 1–6 (2002).
MeSH terms
Substances
Grants and funding
- 310030_205087/Schweizerischer Nationalfonds zur Förderung der Wissenschaftlichen Forschung (Swiss National Science Foundation)
- PP00P3_187198/Schweizerischer Nationalfonds zur Förderung der Wissenschaftlichen Forschung (Swiss National Science Foundation)
- 310030_205087/Schweizerischer Nationalfonds zur Förderung der Wissenschaftlichen Forschung (Swiss National Science Foundation)
- 715289/EC | EU Framework Programme for Research and Innovation H2020 | H2020 Priority Excellent Science | H2020 European Research Council (H2020 Excellent Science - European Research Council)
- MB22.00075/Federal Department of Economic Affairs, Education and Research, Switzerland | Staatssekretariat für Bildung, Forschung und Innovation (State Secretariat for Education, Research and Innovation)
LinkOut - more resources
Full Text Sources

