The origin of modern North Africans as depicted by a massive survey of mitogenomes
- PMID: 40707575
- PMCID: PMC12289972
- DOI: 10.1038/s41598-025-12209-x
The origin of modern North Africans as depicted by a massive survey of mitogenomes
Abstract
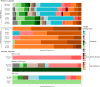
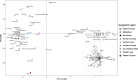
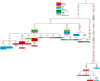
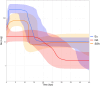
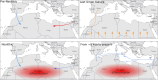
North Africa occupies a unique geographical position in the Mediterranean basin. The genetic profile of modern North African populations is the result of ancient and recent movements of people and periods of isolation. In this paper, we provide a comprehensive mitogenome analysis based on unbiased population sampling from across North Africa including a dataset of 733 modern (238 of which new) and 43 ancient mitogenomes from Morocco, Algeria, Tunisia, Libya and Egypt, compared with modern mitogenomes from Eurasia and sub-Saharan Africa. Our analyses, based on a phylogeographic and demographic approach suggest that mitogenome variability in North African populations (i) can be traced back to three main origins - Eurasia, North Africa and sub-Saharan Africa - and (ii) has been defined during crucial key time periods - from the Palaeolithic to the last millennia. In particular, we identified a signal for population movements from sub-Saharan Africa to North Africa during the favourable climatic period of the Green Sahara, providing evidence from complete mitogenomes that the human dispersals associated with this period also involved females.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests.
Figures
References
-
- Hublin, J. J. et al. New fossils from Jebel irhoud, Morocco and the pan-African origin of Homo sapiens. Nature546, 289–292 (2017). - PubMed
-
- Hunt, C. et al. Site formation processes in caves: the holocene sediments of the Haua fteah,Cyrenaica, Libya. J. Archaeol. Sci.37, 1600–1611 (2010).
-
- Barton, R. N. E. et al. Origins of the iberomaurusian in NW africa: new AMS radiocarbon dating of the middle and later stone age deposits at Taforalt cave, Morocco. J. Hum. Evol.65, 266–281 (2013). - PubMed
-
- Cavalli-Sforza, L. L., Cavalli-Sforza, L., Menozzi, P. & Piazza, A. The History and Geography of Human Genes (Princeton University Press, 1994).
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

