Simultaneous expression of epithelial and immune cell markers in circulating tumor cells identified in patients with stage 4 breast cancer
- PMID: 40707771
- PMCID: PMC12290075
- DOI: 10.1038/s43856-025-01024-0
Simultaneous expression of epithelial and immune cell markers in circulating tumor cells identified in patients with stage 4 breast cancer
Abstract
Background: Heterogeneous circulating tumor cells (CTCs) have been implicated in the formation of new metastases. However, circulating cells expressing both tumor and immune cell proteins are often dismissed as insignificant findings in CTC studies.
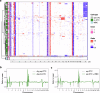
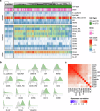
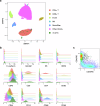
Methods: Two non-contemporaneous blood samples from a metastatic breast cancer patient were analyzed using an enrichment-free platform to identify canonical, epithelial-only CTCs (CD45-/cytokeratin + , epi.CTCs) and CD45 + /cytokeratin+ immune-like CTCs (im.CTCs). Single cells from both samples were subjected to copy number and protein expression profiling. A cohort of 36 metastatic breast cancer patients was then analyzed to search for additional cases with im.CTCs.
Results: Here, we identified and characterized a population of CTCs exhibiting an immune-like state. In two samples from an index patient, im.CTCs outnumbered epi.CTCs, comprising >97% of the CTC population. Single-cell copy number analysis of 43 im.CTCs and 30 epi.CTCs revealed clonal alterations across both populations, confirming a shared tumor origin. Furthermore, im.CTCs contained pseudo-diploid profiles that did not reflect dilution from the addition of a normal diploid genome, indicating that they were unlikely to have originated from tumor-immune cell fusion. Protein expression analysis showed that im.CTCs express CD45 as well as other immune-related markers, such as CD3 and CD4, and the cancer stemness marker, CD44. Subsequent analysis of a metastatic breast cancer cohort identified an additional patient harboring im.CTCs with the same tumor-derived, non-fusion genome as in the index case.
Conclusions: Collectively, these genomic and proteomic features distinguish im.CTCs from previously reported circulating cells may represent a novel form of tumor cell plasticity.
Plain language summary
Tumor cells are known to take on features that allow them to survive and move to new sites. This variation can make it difficult to distinguish them from other cells in the blood. Using a platform to profile rare cells in blood samples, we identified a population of cells expressing cancer and immune cell proteins in a breast cancer patient. Genomics data confirmed that these cells originated from the tumor and that they were different from another cell type sharing a similar protein expression pattern. We analyzed additional samples and found a second patient with these immune-like tumor cells. These findings support the existence of a cancer-immune state that might play a role in helping tumor cells spread.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: P.K. and J.H. hold and receive royalties from Epic Sciences for licensed technology. Epic Sciences and the University of Southern California, USC Michelson Center (P.K. and J.H.), have signed a sponsored research agreement to advance next-generation liquid biopsy technology for precision oncology. All other authors have no conflicts to report.
Figures



References
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

