Interpretable machine learning-guided single-cell mapping deciphers multi-lineage pancreatic dysregulation in type 2 diabetes
- PMID: 40707916
- PMCID: PMC12291316
- DOI: 10.1186/s12933-025-02865-8
Interpretable machine learning-guided single-cell mapping deciphers multi-lineage pancreatic dysregulation in type 2 diabetes
Abstract
Background: Pancreatic cellular heterogeneity is fundamental to systemic metabolic regulation, yet its pathological remodeling in diabetes remains poorly characterized.
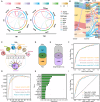
Methods: We integrated single-cell RNA sequencing with machine learning frameworks to decode pancreatic heterogeneity. Novel tools included PanSubPred (two-stage feature selection/XGBoost classifier) for multi-lineage annotation and PSC-Stat (XGBoost/Gini optimization) for stellate cell activation analysis.
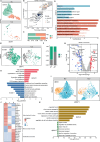
Results: By establishing PanSubPred, we systematically decoded pancreatic cellular diversity, identifying 64 cell-type-specific markers (38 novel) that maintained cross-dataset accuracy (AUC > 0.970) even after excluding known canonical markers. Building on this annotation precision, we developed PSC-Stat to quantify stellate cell activation dynamics, revealing their progressive activation from diabetes to pancreatic cancer (activated/quiescent ratio: control: 1.44 ± 1.02, diabetes: 4.72 ± 4.01, pancreatic cancer: 18.67 ± 18.70). Diabetes reorganized intercellular communication into ductal-centric hubs via FGF7-FGFR2/3, EFNB3-EPHB2/4/6 and EFNA5-EPHA2 axes, from which we derived a 15-gene signature for diabetic ductal cells (AUC = 0.846). Beta cell heterogeneity analysis uncovered diabetes-associated depletion of mature insulin-secretory clusters (INS + NKX6-1+), expansion of immature (CD81 + RBP4+) and endoplasmic reticulum stress-adapted subtypes (DDIT3 + HSPA5+). Moreover, non-beta lineages exhibited parallel dysfunction: acinar cells shifted toward inflammatory states (CCL2 + CXCL17+), while ductal cells adopted secretory phenotypes (MUC1 + CFTR+).
Conclusions: This study presents a machine learning-based single-cell framework that systematically maps pancreatic cellular alterations in diabetes. The identified novel signatures, stellate activation dynamics, and beta cell maturation trajectories may serve as potential targets for diabetic management and pancreatic cancer risk stratification.
Keywords: Beta cell dysfunction; Machine learning; Pancreatic cellular heterogeneity; Single-cell transcriptomics; Stellate cell activation; Type 2 diabetes.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Not applicable. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures



References
-
- Saltiel AR, Kahn CR. Insulin signalling and the regulation of glucose and lipid metabolism. Nature. 2001;414(6865):799–806. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

